
Dr Lachlan Jolly
Senior Research Fellow
School of Pharmacy and Biomedical Sciences
College of Health
Eligible to supervise Masters and PhD - email supervisor to discuss availability.
Dr Jolly is head of the Neurobiology Research Group and investigates the genetic, molecular, cell and developmental processes which underpin brain development. The overarching goal is to provide patients with neurodevelopmental disorders (NDDs) clear treatment options tailored to underlying genetic mechanisms. The key aspects are therefore to (1) Identify the underlying genetics causes, and (2) Discover tailored therapies. The bridge between these two aspects requires an understanding of how the genes involved in NDDs actually control the development of the brains. The major research areas are to develop technologies to discover causative mutations, apply patient, stem and neuronal cell of brain development to understand gene function, and discover new therapeutics and biomarkers.
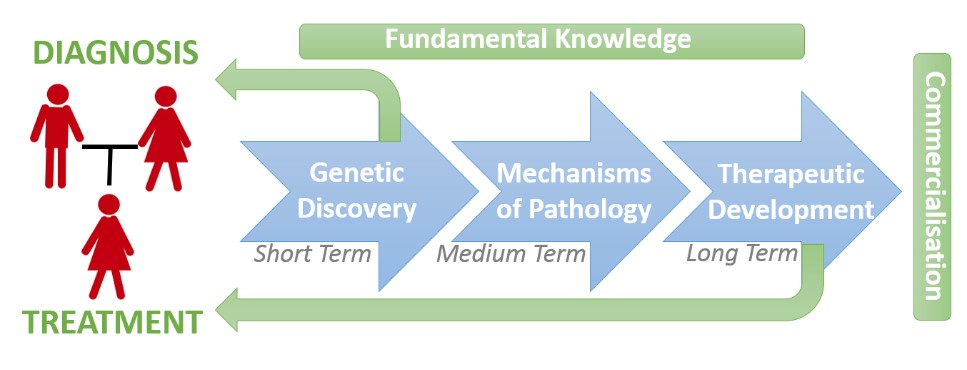
Neurodevelopmental Disorders (NDDs) including e.g. intellectual disability, autism spectrum disorder, and movement disorders among others affect ~2% of the world’s population. In developed countries, genetic mutation is the common cause, and cannot always be prevented. With ~1000 genes, and 10,000s of variants implicated in NDDs, understanding each gene/variant function is a major impediment to developing personalised medicines. Our strategy is to target ‘points of convergence’ that exist between NDDs of different genetic origin and study them to identify and develop new therapeutic approaches. We have discovered and studied the involvement of cellular degradation pathways in NDDs which serve as molecular points if convergence. These pathways include both Non-sense Mediated mRNA Decay Pathway (NMD) which regulates mRNA degradation, and protein ubiquitylation which regulates protein degradation. Study and manipulation of these pathways has the potential to inform and influence brain pathologies of NDDs from diverse genetic origins.
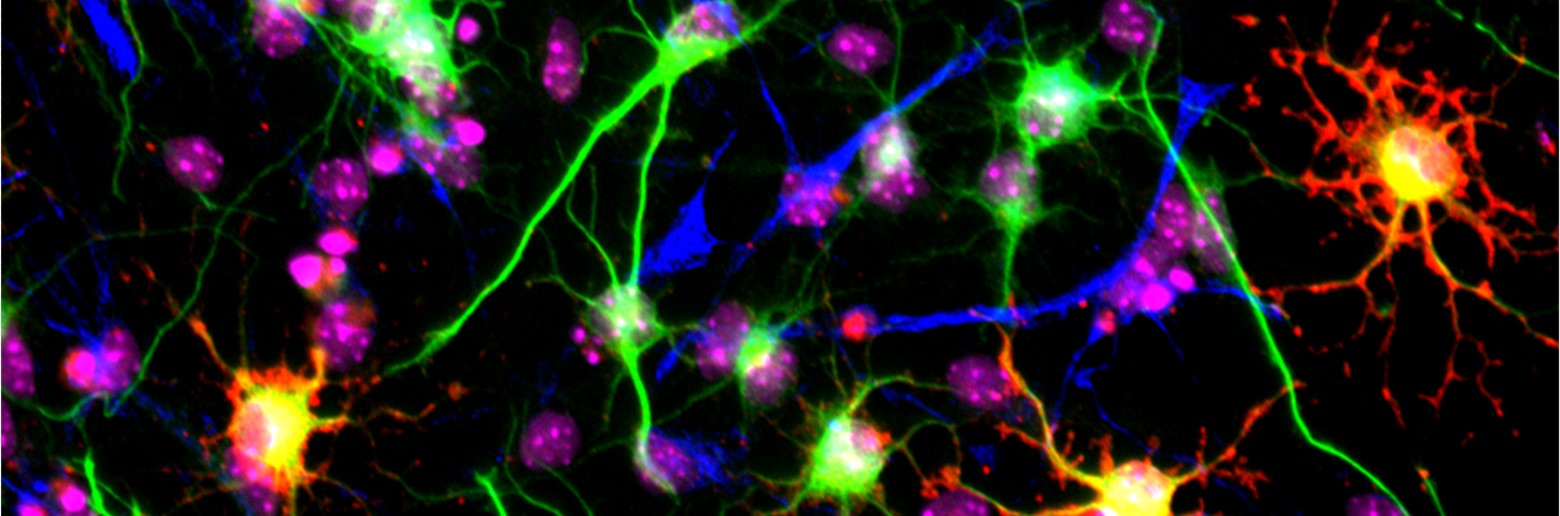
Project 1 Nonsense Mediated mRNA Decay: Dr Jolly discovered that mutations in genes encoding the core factors of the Nonsense Mediated mRNA Decay (NMD) factors cause neurodevelopmental disorders. He pioneered how NMD controls stem cell and neuronal cell function. Dr Jolly now leads projects funded by NHMRC Ideas Grant (2021-24) and Sanfilippo Foundation (2021-2022) to investigate and harness the NMD pathway to treat genetic disease caused by nonsense mutations which cause 13% of all genetic diseases.
This research aims to understand the role of NMD during brain development and predict & alleviate outcomes of diseases caused by nonsense mutations. It is underpinned by the development tools to quantify NMD using human biomarkers and engineered biosensors to permit tracking of the NMD pathway in cells and organisms and aid the identification of drugs molecules to modify NMD.
Aim 1. Study the impact of compromised NMD function on brain development.
Aim 2. Use live-cell NMD biosensors to track NMD during development.
Aim 3. Identify NMD biomarkers that predict disease severity and responsiveness to therapies.
Aim 4. Identify FDA approved NMD inhibitory molecules.
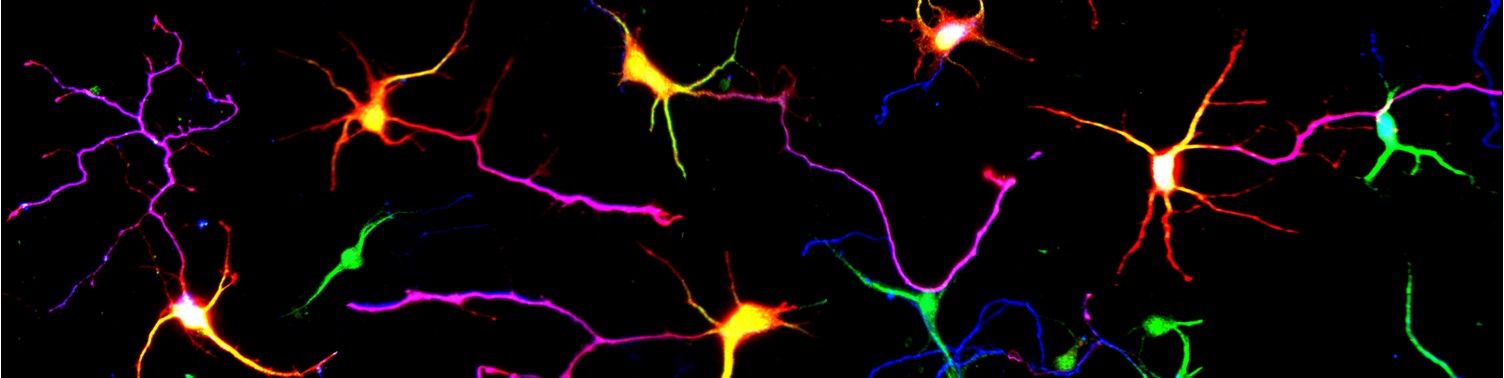
Project 2: Protein Degradation: Dr Jolly has been investigating ubiquitin dependent protein degradation by understanding the process of protein de-ubiquitination in the context of brain development. He has >20 years expertise in the field. He has defined clinical syndromes associated with defective de-ubiquitination, developed tools to help diagnose patients, established a global collaborative network of clinicians and scientists, established a patient support group, and discovered how protein deubiquitination is essential for stem and neuronal cell function. His research is translating this knowledge to overcome a common genetic mechanism known as haploinsufficiency, which relevant to ~3000 genetic diseases, and driven by loss of protein dosage.
This research investigates how loss protein dosage can be controlled ubiquitin dependent degradation during brain development and homeostasis.
Aim 1. Identify brain protein networks whose abundance is controlled by deubiquitinating enzymes.
Aim 2. Identify mechanisms that activate deubiquitinating enzymes
Aim 3. Antagonise the ubiquitin proteasome system to overcome effects of haploinsufficiency.
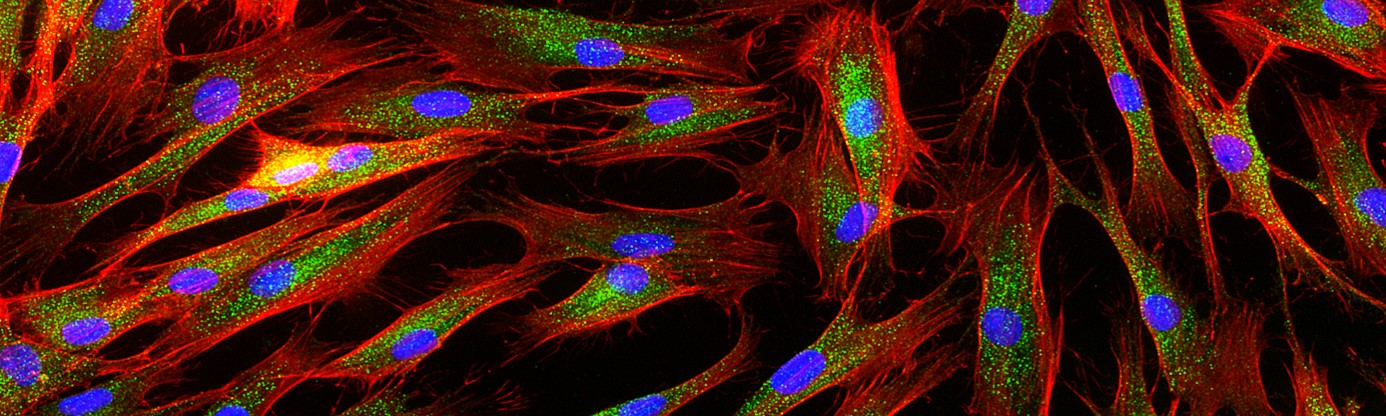
Project 3: Discovery of disease causing splice altering variants. To date, more than 2000 genes are implicated in Neurodevelopmental Disorders (NDDs). Whilst many different types of variants can cause detriment to NDD gene function, those impacting mRNA splicing and processing are both among the most difficult to identify from DNA sequence alone, and have great potential to be ‘treatable’ through development of new generation antisense oligo therapies among others. Indeed, many NDD patients go undiagnosed as the molecular action of the gene splicing variants cannot be accurately assessed due to the inability to test the variant(s) effect in the context of the living cell of the patient. Close to 500 NDD genes cannot be robustly assayed in patient-derived blood and skin cells because they are simply not expressed. We developed a technology based on modified CRISPR dCas9 gene editing that enables the study of silent genes in patient-derived cells. We will develop and apply this technology to provide new diagnostic avenues including to evaluate the molecular mechanisms of variants that alter mRNA processing and/or degradation of silent NDD genes.
Aim 1: Identify the silent neurodevelopmental disorder genes conducive to dCas9 transactivation.
Aim 2. Identify the silent neurodevelopmental genes expressed in neurons derived by transdifferentiation.
Aim 3. Analyse the pathogenicity of splicing variants in neurodevelopmental disorder patient cell lines.
| Date | Position | Institution name |
|---|---|---|
| 2022 - 2026 | First 1000 Days Fellow | University of Adelaide |
| 2020 - 2020 | Robinson Research Institute Career Development Fellow | University of Adelaide |
| 2016 - 2018 | Discovery Early Career Research Fellow | Australian Research Council |
| 2015 - 2015 | Senior Lecturer | University of Adelaide |
| Date | Type | Title | Institution Name | Country | Amount |
|---|---|---|---|---|---|
| 2019 | Award | AFGN Meeting Best Post-doc Talk | Australian Functional Genomics Network | Australia | - |
| 2016 | Fellowship | Discovery Early Career Research Award | Australian Research Council | Australia | 378000 |
| 2013 | Award | Adelaide Protein Group Early Career Award | Australian Society for Biochemistry and Molecular Biology | Australia | - |
| 2012 | Award | Toshiya Yamada Early Career Award | Australian and New Zealand Society for Cell and Developmental Biology | Australia | - |
| Date | Institution name | Country | Title |
|---|---|---|---|
| 2005 - 2010 | University of Adelaide | Australia | PhD |
| Date | Title | Institution | Country |
|---|---|---|---|
| 2013 - 2013 | Research Placement | University of California, San Diego | United States |
| Year | Citation |
|---|---|
| 2015 | Shoubridge, C., Hinze, S., Lie, S., & Jolly, L. (2015). Functional deficit of IQSEC2, a known intellectual disability gene, disrupts normal dendritic spine morphogenesis. In JOURNAL OF NEUROCHEMISTRY Vol. 134 (pp. 308-309). Cairns, AUSTRALIA: WILEY-BLACKWELL. |
| 2005 | Jolly, L. A., & Wood, S. A. (2005). The role of Fam in murine neurogenesis. In MECHANISMS OF DEVELOPMENT Vol. 122 (pp. S180). ELSEVIER SCIENCE BV. |
Successful applications I have been involved in have received in total >$2 million dollars. This includes (Category1) $378K as ARC DECRA , $764K as CIB on NHMRC Project Grant, $238K as CIC on ARC Discovery Grant; and (Category II) $325K as CIA across 5x NFP Grants.
- (2010) Chief Investigator A. Women’s and Children’s Hospital Foundation Grant. $50 K. Title “Modelling the molecular pathogenesis of intellectual disability: Functional importance of the nonsense mediated mRNA decay (NMD) factor UPF3B”.
- (2011) Chief Investigator A. Women’s and Children’s Hospital Foundation Grant. $50 K. Title “Investigating the cell and molecular neurobiology of Usp9x: resolving the pathological mechanisms behind the genetic causes of intellectual disability”.
- (2013) Chief Investigator A. Women’s and Children’s Hospital Foundation Grant. $75K. Title “Molecular pathology of HCFC1, a novel modifier of embryonic neural cell behaviour involved in intellectual disability”.
- (2014) Chief Investigator A. Women’s and Children’s Hospital Foundation Grant. $75 K. Title “Rescue of intellectual disability severity through modification of the non-sense mediated mRNA decay factor UPF3A”.
- (2014) Chief Investigator B. NH&MRC Project Grant: 1063808. $764 K. Title “The role of UPF3B and nonsense mediated mRNA decay surveillance in the pathology of intellectual disability”.
- (2015) Chief Investigator A. Channel 7 Children’s Foundation Grant. $75 K. Title “Visualising Nonsense Mediated mRNA Decay”.
- (2015) Chief Investigator B. RRI Innovation Seed Funding Program. $30 K. Title “CRISP-up: a novel Crispr/Cas9 based approach to gene overexpression”.
- (2016) Chief Investigator A. RRI Innovation Seed Funding Program. $25 K. Title “Generation of Pluripotent Stem Models for the study of Human Neurodevelopmental Disorders”.
- (2016) Chief Investigator C. RRI Innovation Seed Funding Program. $25 K. Title “The impact of Zika virus infection on placenta and neural development during early gestation”.
- (2017) Chief Investigator C. ARC Discovery Grant: DP170103090. $234 K. Title “TREX-mediated mRNA export: critical pathways in differentiation and function”.
- (2017) Co-PI. Penn Medicine Orphan Disease Centre. $70K. Titled “Understanding neurogenesis in MPS IIIA disease”.
- (2017) Chief Investigator C. SAFRI Simons Autism Funds. $100K. Title “USP9X in Autism”
- (2018) Chief Investigator A. RRI Invest for Success Funding Program. 20K. Title “Control of postnatal hippocampal development by intellectual disability genes”.
- (2019) Chief Investigator A. RRI Invest for Success Funding Program. 30K. Title “Control of postnatal hippocampal development by intellectual disability genes”.
Fellowships
- (2016-2018) ARC Discovery Early Career Research Award (DECRA): DE160100620. $378 K. Title “Seeing through the nonsense: Gene regulation and the nonsense mediated mRNA decay pathway”.
- (2020) Robinson Research Institute Career Development Fellowship. $100K. Title “Cellular Degradation Pathways In Neurodevelopmental Disorders”
HDR Students
- (2012-18). PhD. Dr Claire Homan, University of Adelaide.
- (2016-18). PhD. Dr Rebecca Lehmann, University of Adelaide.
- (2014). Undergraduate placement. Ms Deepti Domingo, University of Adelaide.
- (2013-20). PhD. Dr Debrah Renders, University of Adelaide.
- (2015-20). PhD. Dr Deepti Domingo, University of Adelaide.
- (2020-21). Masters. Ms Emmylou Nicolas.
Undergraduate Lectures
- (2016-current) University of Adelaide: Biomedical Science II, Adelaide, Australia
- (2018-current) University of Adelaide: Cellular Neuroscience III, Adelaide, Australia
| Date | Role | Research Topic | Program | Degree Type | Student Load | Student Name |
|---|---|---|---|---|---|---|
| 2025 | Principal Supervisor | Functional characterisation of enhancer variants associated with common epilepsy | Doctor of Philosophy | Doctorate | Full Time | Miss Chano Ranwala |
| 2025 | Principal Supervisor | Role of RNA processing in Human Development and Rare Genetic Diseases | Doctor of Philosophy | Doctorate | Full Time | Miss Emmylou Nicolas |
| 2025 | Principal Supervisor | Functional characterisation of enhancer variants associated with common epilepsy | Doctor of Philosophy | Doctorate | Full Time | Miss Chano Ranwala |
| 2025 | Principal Supervisor | Role of RNA processing in Human Development and Rare Genetic Diseases | Doctor of Philosophy | Doctorate | Full Time | Miss Emmylou Cortez Nicolas-Martinez |
| 2024 | Principal Supervisor | Assessing the inaccessible: Application of direct cellular reprogramming and genome editing techniques to address current diagnostic limitations in Neurodevelopmental Disorders. | Doctor of Philosophy | Doctorate | Full Time | Ms Olivia Robinson |
| 2024 | Principal Supervisor | Long non-coding RNAs as regulators of neuronal differentiation and neuroblastoma | Doctor of Philosophy | Doctorate | Full Time | Miss Regina Belen Peralta Callanta |
| 2024 | Principal Supervisor | The Pathological Mechanisms Underpinning USP9X Syndrome | Doctor of Philosophy | Doctorate | Full Time | Miss Charlotte Yu |
| 2024 | Co-Supervisor | Identification and Characterization of Genetic Determinants of Neurodevelopment | Doctor of Philosophy | Doctorate | Full Time | Miss Saadia Maryam Saadi |
| 2024 | Principal Supervisor | Assessing the inaccessible: Application of direct cellular reprogramming and genome editing techniques to address current diagnostic limitations in Neurodevelopmental Disorders. | Doctor of Philosophy | Doctorate | Full Time | Ms Olivia Robinson |
| 2024 | Principal Supervisor | The Pathological Mechanisms Underpinning USP9X Syndrome | Doctor of Philosophy | Doctorate | Full Time | Miss Charlotte Yu |
| 2024 | Co-Supervisor | Identification and Characterization of Genetic Determinants of Neurodevelopment | Doctor of Philosophy | Doctorate | Full Time | Miss Saadia Maryam Saadi |
| 2024 | Principal Supervisor | Long non-coding RNAs as regulators of neuronal differentiation and neuroblastoma | Doctor of Philosophy | Doctorate | Full Time | Miss Regina Belen Peralta Callanta |
| Date | Role | Research Topic | Program | Degree Type | Student Load | Student Name |
|---|---|---|---|---|---|---|
| 2023 - 2025 | Co-Supervisor | Harnessing transcriptomic data to better understand gene regulatory mechanisms of neurodevelopmental disorders | Doctor of Philosophy | Doctorate | Full Time | Ms Urwah Nawaz |
| 2016 - 2019 | Co-Supervisor | The Neurogenic Potential of Stem Cells is Altered in Mucopolysaccharidosis Type IIIA | Doctor of Philosophy | Doctorate | Full Time | Miss Rebecca Jayne Lehmann |
| 2016 - 2020 | Co-Supervisor | A Study of Nonsense Mediated mRNA Decay using Naturally Occurring Genetic Variants and Through the Development of a Synthetic Transgene | Doctor of Philosophy | Doctorate | Full Time | Miss Dee Dianna Domingo |
| 2014 - 2020 | Co-Supervisor | The Role of UPF3A and UPF3B in Early Development and Neural Differentiation | Doctor of Philosophy | Doctorate | Full Time | Ms Debrah Sadie Sebolai |
| 2012 - 2017 | Co-Supervisor | Neurobiology of PCDHI9-Female Epilepsy | Doctor of Philosophy | Doctorate | Full Time | Dr Claire Homan |







