
Professor Jose Polo
Director of Adelaide Centre for Epigenetics
School of Biomedicine
Faculty of Health and Medical Sciences
Eligible to supervise Masters and PhD - email supervisor to discuss availability.
Jose Maria Polo was born in Buenos Aires, Argentina where he graduated from Buenos Aires University as a Biochemist. In 2002, Jose began his graduate studies at Albert Einstein College of Medicine, New York under the supervision of Dr. Ari Melnick where he worked on the transcriptional mechanism of the BCL6 repression complex in B-cell lymphomas and B-cell maturation. In 2008, he obtained his PhD and moved to Boston to the laboratory of Dr. Konrad Hochedlinger at the Harvard Stem Cell Institute to work on reprogramming of adult cells into induced pluripotent stem (iPS) cells. In particular, his work focused on the acquisition of immortality and the existence of epigenetic memory during reprogramming. In June 2011, established his independent research group at Monash University, where he holded appointments to the departments of Anatomy and Developmental Biology and to the Australian Regenerative Medicine Institute. In 2012, Jose was awarded a NHMRC Career Development Fellowship, in 2014 a Silvia and Charles Viertel Senior Medical Research Fellowship and in 2018 a Future Fellowship to continue his work in the molecular mechanism governing the reprogramming process and the epigenetic mechanism underpinning cell fate. In October 2021, Jose was recruited to the University of Adelaide as the inaugural Director of the Adelaide Centre for Epigenetics (ACE) and group leader of the recently established South Australian Immunogenomics Cancer Institute (SAiGENCI). In Adelaide, he will continue his work in epigenetics and its application to reprogramming, early embryogenesis and cancer. His work in epigenetics, reprogramming and cancer has been published in journals such as Nature, Cell, Nature Genetics, Cell Stem Cell and Nature Medicine among others as well as recognised with several awards including the Merit Award from the American Society of Haematology, the inaugural Metcalf Award, Victorian Young Tall Poppy Award, the Monash’s Vice-Chancellor award. In 2016, he co-founded Mogrify Ltd to translate reprogramming technologies into therapies, receiving several accolades including the 2019 Scrip Innovation Award.
- My Research
- Career
- Publications
- Grants and Funding
- Teaching
- Supervision
- Professional Activities
- Contact
My Research
The Polo group is interested in the transcriptional and epigenetic mechanisms that govern cell identity, in particular pluripotency, reprogramming of somatic cells into induced pluripotent stem (iPS) cells, development and cancer.
Being able to reprogram any specific mature cellular program into a pluripotent state and from there back into any other particular cellular program provides a unique tool to dissect the molecular and cellular events that permit the conversion of one cell type to another. Moreover, iPS cells and the reprogramming technology are of great interest in pharmaceutical and clinical settings, since the technology can be used to generate animal and cellular models for the study of various diseases as well in the future to provide specific patient tailor made cells for their use in cellular replacement therapies. By using a broad array of approaches through the use of mouse and human stem cells models combined with different molecular, biochemical, cellular techniques and genome wide approaches, our lab will aim to dissect the nature and dynamics of such events. Understanding the events leading to the generation of iPS cells has allowed us to not only she light into this process but it has led to the generation of complex models of humans development. Furthermore, we are using and translating the tools and knowledge gained through this work into the fields of development, cancer and several other diseases.
We are particularly interested in the following aspects:
1) The kinetics and universality of the epigenetic and genomic changes occurring during cell fate transitions.
2) The composition and assembly kinetics of transcriptional regulation complexes at pluripotency genes and cancer associated transcription factors.
3) How the cell of origin influences the in vitro and in vivo plasticity potential of cells generated during the reprogramming process.
4) The role of transcription factors in cancer initiation and progression
5) Understanding mammalian development through the use of complex in vitro models
6) Synthetic cell biology

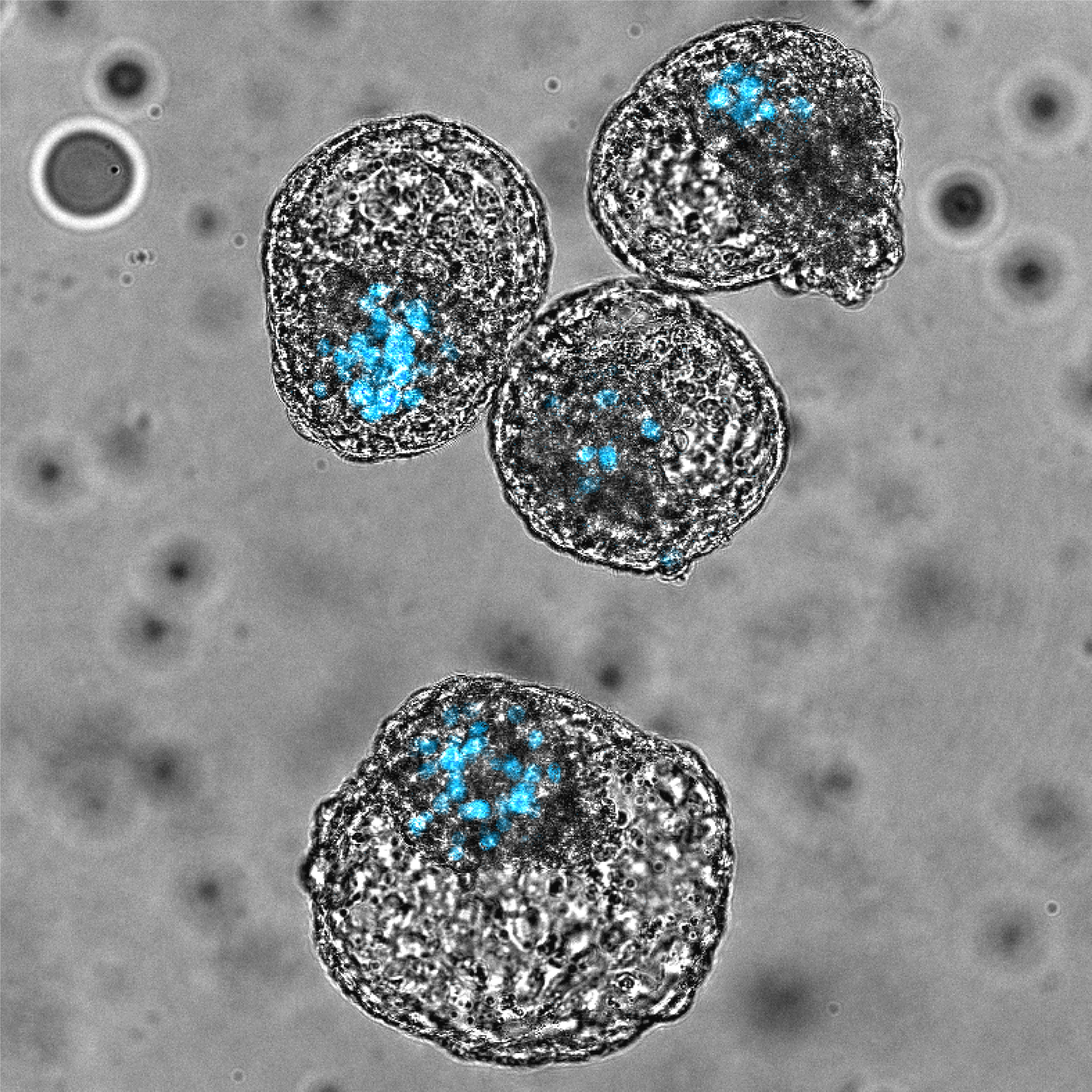
Higher Degree Research Projects
Understanding the epigenetic and transcriptional changes during reprogramming of cells to pluripotent stem cells.
The derivation of human embryonic stem cells (hESCs) and, more remarkably, the generation of human induced pluripotent stem cells (iPSCs) has revolutionised our understanding of pluripotency and opened new avenues for disease modelling, drug screening and regenerative medicine. The ability to reprogram any mature cell back to a pluripotent state, and then into another cell type, provides a unique opportunity to dissect the molecular and cellular events that occur during this conversion. Our lab aims to understand the kinetics and universality of the epigenetic and genomic changes occurring during reprogramming, the composition and assembly kinetics of transcriptional regulation complexes of pluripotency genes and how the cell of origin influences the in vitro and in vivo plasticity potential of cells generated during the reprogramming process. We will achieve this by combining stem cell technologies with several different molecular, biochemical, cellular techniques and genome-wide approaches; including ChIP, CUT&RUN, and single cell “omics”.
Molecular investigation of induced trophoblast cells (iTSC) and their derivatives.
The derivation of human embryonic stem cells (hESCs) and, more remarkably, the generation of human induced pluripotent stem cells (iPSCs) has revolutionised our understanding of pluripotency and opened new avenues for disease modelling, drug screening and regenerative medicine. However, the trophectoderm (TE) gives rise to the placenta and as such, iPSCs cannot provide models for TE or placenta. To address this important challenge, our lab has generated iTSCs for the first time. Our lab will use this model with both human and non-human primate cells, to understand the transcriptional and epigenomic changes that occur in vitro in the early human trophoblast and demonstrate that iTSCs can be used for disease modelling.
Pluripotency factors and cancer
Pluripotent stem cells can self-renew indefinitely and give rise to all cells of the adult organism. These remarkable capacities are the result of a core transcriptional network controlled by OCT4, SOX2 and NANOG, whose expression is lost upon differentiation. Several cancers have been shown to reactivate OCT4, SOX2 and/or NANOG, with high expression levels positively correlating with cancer progression and severity. Since they are linked to proliferative and multi-lineage differentiation capacity of cancer cells, they present attractive anticancer targets. However, they are considered “undruggable” since they lack catalytic active sites for molecules to bind.
To provide therapeutic alternatives, the Polo lab has adapted and developed various novel techniques to determine how expression and function of the pluripotency factors OCT4, SOX2 and NANOG are controlled in various physiological and pathological cell types, including embryonic stem cells and cancer respectively.
Uncovering the regulatory complex of Bcl6 in lymphomas
Cellular identity is controlled by transcription factors, which bind to specific regulatory elements (REs) within the genome to regulate gene expression and cell fate changes. Recent advances in epigenome profiling techniques have significantly increased our understanding of which REs are utilised in which cell type, however, which factors interact with these REs remains largely elusive. A major impediment to dissecting protein complexes at specific genomic loci is the shortage of appropriate techniques. The most common technique to assess TF binding is chromatin immunoprecipitation (ChIP), which relies on antibodies to interrogate the binding sites of a single TF. Yet, ChIP does not allow dissection of the composition of a multi-protein complex at a specific locus. Importantly, the Polo lab has developed a novel epigenetic technique termed TINC (TALE-mediated Isolation of Native Chromatin), which allows us to do exactly that (Knaupp et al., Stem Cell Reports 2020). TINC relies on epitope-tagged TALEs, which are DNA-binding proteins engineerable to target specific genomic regions. Upon cross-linking of the cells, the target regions are isolated based on affinity purification of the TALE and associated nucleic acid and protein molecules are analysed by next generation sequencing and mass spectrometry, respectively. In our proof-of-concept experiments, we dissected the protein complex formed at the Nanog promoter, a key pluripotency RE. We identified TFs previously known to bind to this locus as well as novel proteins whose role in pluripotency we further validated (Knaupp et al., Stem Cell Reports 2020). Consequently, with this valuable technique at hands, this PhD project aims at deciphering how the BTB/POZ transcriptional repressor and oncogene BCL6 is (mis)regulated in B-cell lymphomas, which in turn has major potential in identifying novel therapeutic targets.
-
Appointments
Date Position Institution name 2021 - ongoing Professor of Epigenetics and Director of the Adelaide Centre for Epigenetics University of Adelaide 2018 - 2021 Unit coordinator for Master of Biotechnology Australian Regenerative Medicine Institute 2011 - ongoing Professor and Group Leader in the Faculty of Medicine, Nursing and Health Sciences Monash University 2011 - 2021 Dual appointment with the Australian Regenerative Medicine Institute (ARMI) and the Department of Anatomy and Developmental Biology (ADB) Monash University 2008 - 2011 Research Fellow Harvard Stem Cell Institute and Centre for Regenerative Medicine, 2002 - 2008 Graduate Student Albert Einstein College of Medicine 2000 - 2002 Junior Lecturer University of Buenos Aires 1998 - ongoing Visiting Student Memorial Sloan Kettering Cancer Center 1995 - 2002 Research Assistant University of Buenos Aires 1995 - 2000 Teaching Assistant University of Buenos Aires -
Awards and Achievements
Date Type Title Institution Name Country Amount 2019 Award MSD’s Innovation Award at the 15th Annual Scrip Awards Mogrify Ltd United Kingdom - 2018 Nomination Selected as Next Generation of Leaders of the International Society for Stem Cell Research International Society for Stem Cell Research Australia - 2018 Fellowship ARC Future Fellowship ARC Australia - 2017 Award Vice-Chancellor’s Diversity and Inclusion Award Monash University Australia - 2015 Achievement Specialist Plenary Speaker, Australian Academy of Science’s Theo Murphy High-Flyer Think Tank, Australia Australian Academy of Science’s Theo Murphy High-Flyer Think Tank Australia - 2014 Award Metcalf Award National Stem Cell Foundation of Australia Australia - 2014 Achievement Invited as one of eight promising early career scientists (from all scientific disciplines across the world) of the Future Leaders program of the Science and Technology in Society Forum in Kyoto Future Leaders program of the Science and Technology in Society Forum in Kyoto Australia - 2014 Award Deans Award for Excellence in Research (Early Career) Monash University Australia - 2014 Fellowship Sylvia and Charles Viertel Senior Medical Research Fellowship Sylvia and Charles Viertel Australia - 2013 Award Victorian Young Tall Poppy Science Award Victorian Young Tall Poppy Science Australia - 2012 Fellowship Career Development Fellowship - NHRMC, Australia NHMRC Australia - 2011 Fellowship Larkins Fellowship Monash Universit Australia - 2009 Award Postdoctoral Fellowship Award Massachusetts General Hospital United States - 2005 Award Pre-doctoral Fellowship National Cancer Centre Australia - 2004 Award Merit Award and Plenary Speaker from the American Society of Haematology American Society of Haematology United States - -
Education
Date Institution name Country Title 2002 - 2008 Albert Einstein College of Medicine United States Doctor of Philosophy (PhD) 2002 - 2004 Albert Einstein College of Medicine United States Master of Science (MSci) 1993 - 2000 University of Buenos Aires Argentina Biochemist (Professional Degree)
-
Journals
Year Citation 2025 Kan, W. L., Weekley, C. M., Nero, T. L., Hercus, T. R., Yip, K. H., Tumes, D. J., . . . Parker, M. W. (2025). The β Common Cytokine Receptor Family Reveals New Functional Paradigms From Structural Complexities. Immunological Reviews, 329(1), e13430-1-e13430-25.
Scopus1 WoS3 Europe PMC12025 Vandenbempt, V., Roach, M., Mohenska, M., Wise, K., Wang, T., Harvey, K., . . . Martelotto, L. G. (2025). Validation of single-cell transcriptomic profiles from Illumina and MGI Tech sequencing platforms. FEBS Letters, 599(17), 2533-2542.
2025 Pitino, E., Pascual-Reguant, A., Segato-Dezem, F., Wise, K., Salvador-Martinez, I., Crowell, H. L., . . . Martelotto, L. G. (2025). STAMP: Single-cell transcriptomics analysis and multimodal profiling through imaging.. Cell, 188(18), 5100-5117.e26.
Scopus1 Europe PMC12025 Bienroth, D., Charitakis, N., Wong, D., Zhang, Y. C., Jaeger-Honz, S., Ding, J., . . . Ramialison, M. (2025). Automated integration of multi-slice spatial transcriptomics data in 2D and 3D using VR-Omics. Genome Biology, 26(1), 182.
Europe PMC12025 Kroeger, B., Manning, S. A., Mohan, V., Lou, J., Sun, G., Lamont, S., . . . Harvey, K. F. (2025). Hippo signaling regulates the nuclear behavior and DNA binding times of YAP and TEAD to control transcription. Science Advances, 11(30), eadw4974-1-eadw4974-16.
2025 Gartner, M. J., Smith, M. L., Dapat, C., Liaw, Y. W., Tran, T., Suryadinata, R., . . . Neil, J. A. (2025). Contemporary seasonal human coronaviruses display differences in cellular tropism compared to laboratory-adapted reference strains. Journal of Virology, 99(9), e0068425.
2025 Ranuncolo, S. M., Polo, J. M., Dierov, J., Singer, M., Kuo, T., Greally, J., . . . Melnick, A. (2025). Editorial Expression of Concern: Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR(Nature Immunology, 10.1038/ni1478). Nature Immunology, 26(10), 1837.
2024 Short, K. M., Tortelote, G. G., Jones, L. K., Diniz, F., Edgington-Giordano, F., Cullen-McEwen, L. A., . . . El-Dahr, S. S. (2024). The Impact of Low Protein Diet on the Molecular and Cellular Development of the Fetal Kidney.. bioRxiv.
2024 Applewhite, D. A., Bhalla, N., Cabezas-Wallscheid, N., Erez, N., Li, M., Polo, J. M., . . . Wang, X. (2024). Mentoring the next generation of cell biologists.. Nat Cell Biol, 26(1), 15-18.
2024 Healy, E., Zhang, Q., Gail, E. H., Agius, S. C., Sun, G., Bullen, M., . . . Davidovich, C. (2024). The apparent loss of PRC2 chromatin occupancy as an artifact of RNA depletion. Cell Reports, 43(3), 113858-1-113858-12.
Scopus14 WoS14 Europe PMC162024 Esteban, M. A., & Polo, J. M. (2024). Editorial overview: Early embryonic development models: back to the beginning.. Curr Opin Genet Dev, 85, 102175.
2024 Davidson, K. C., Sung, M., Brown, K. D., Contet, J., Belluschi, S., Hamel, R., . . . Parfitt, G. J. (2024). Single nuclei transcriptomics of the in situ human limbal stem cell niche.. Scientific reports, 14(1), 6749.
Scopus6 WoS6 Europe PMC62024 Tan, J. P., Liu, X., & Polo, J. M. (2024). Reprogramming fibroblast into human iBlastoids. Nature Protocols, 19(8), 2298-2316.
Scopus1 WoS2 Europe PMC12024 Yap, K. K., Schröder, J., Gerrand, Y. W., Dobric, A., Kong, A. M., Fox, A. M., . . . Mitchell, G. M. (2024). Liver specification of human iPSC-derived endothelial cells transplanted into mouse liver. JHEP Reports, 6(5), 101023-1-101023-18.
Scopus5 WoS6 Europe PMC62024 Chowdhury, M. M., Zimmerman, S., Leeson, H., Nefzger, C. M., Mar, J. C., Laslett, A., . . . Cooper-White, J. J. (2024). Superior Induced Pluripotent Stem Cell Generation through Phactr3-Driven Mechanomodulation of Both Early and Late Phases of Cell Reprogramming. Biomaterials Research, 28, 0025-1-0025-19.
Scopus4 WoS4 Europe PMC32024 Yang, G. N., Sun, Y. B. Y., Roberts, P. K., Moka, H., Sung, M. K., Gardner-Russell, J., . . . Daniell, M. (2024). Exploring single-cell RNA sequencing as a decision-making tool in the clinical management of Fuchs’ endothelial corneal dystrophy. Progress in Retinal and Eye Research, 102, 101286.
2024 Williamson, A. E., Liyanage, S., Hassanshahi, M., Dona, M. S. I., Toledo-Flores, D., Tran, D. X. A., . . . Psaltis, P. J. (2024). Discovery of an embryonically derived bipotent population of endothelial-macrophage progenitor cells in postnatal aorta. Nature Communications, 15(1), 7097-1-7097-21.
Scopus6 WoS4 Europe PMC52024 Li, Y., Hunter, A., Wakeel, M. M., Sun, G., Lau, R. W. K., Broughton, B. R. S., . . . Samuel, C. S. (2024). The renoprotective efficacy and safety of genetically-engineered human bone marrow-derived mesenchymal stromal cells expressing anti-fibrotic cargo. Stem Cell Research and Therapy, 15(1), 375-1-375-27.
2024 Scoyni, F., Giudice, L., Väänänen, M. A., Downes, N., Korhonen, P., Choo, X. Y., . . . Malm, T. (2024). Alzheimer's disease-induced phagocytic microglia express a specific profile of coding and non-coding RNAs. Alzheimer's and Dementia, 20(2), 954-974.
Scopus11 WoS11 Europe PMC102024 Liu, X., & Polo, J. M. (2024). Human blastoid as an in vitro model of human blastocysts. Current Opinion in Genetics and Development, 84, 102135-1-102135-8.
Scopus10 WoS14 Europe PMC102023 Chen, J., Neil, J. A., Tan, J. P., Rudraraju, R., Mohenska, M., Sun, Y. B. Y., . . . Polo, J. M. (2023). Author Correction: A placental model of SARS-CoV-2 infection reveals ACE2-dependent susceptibility and differentiation impairment in syncytiotrophoblasts.. Nat Cell Biol, 26(2), 305.
2023 Namipashaki, A., Pugsley, K., Liu, X., Abrehart, K., Lim, S. M., Sun, G., . . . Hawi, Z. (2023). Integration of xeno-free single-cell cloning in CRISPR-mediated DNA editing of human iPSCs improves homogeneity and methodological efficiency of cellular disease modeling. Stem Cell Reports, 18(12), 2515-2527.
Scopus1 WoS1 Europe PMC12023 Rudraraju, R., Gartner, M. J., Neil, J. A., Stout, E. S., Chen, J., Needham, E. J., . . . Subbarao, K. (2023). Parallel use of human stem cell lung and heart models provide insights for SARS-CoV-2 treatment. Stem Cell Reports, 18(6), 1308-1324.
Scopus7 WoS7 Europe PMC102023 Chen, J., Neil, J. A., Tan, J. P., Rudraraju, R., Mohenska, M., Sun, Y. B. Y., . . . Polo, J. M. (2023). A placental model of SARS-CoV-2 infection reveals ACE2-dependent susceptibility and differentiation impairment in syncytiotrophoblasts. Nature Cell Biology, 25(8), 1223-1234.
Scopus17 WoS19 Europe PMC192023 Buckberry, S., Liu, X., Poppe, D., Tan, J. P., Sun, G., Chen, J., . . . Lister, R. (2023). Transient naive reprogramming corrects hiPS cells functionally and epigenetically. Nature, 620(7975), 863-872.
Scopus44 WoS43 Europe PMC522023 Zhou, Y., Xie, L., der, J. S., Schuster, I. S., Nakai, M., Sun, G., . . . Mackay, C. R. (2023). Dietary fiber and microbiota metabolite receptors enhance cognition and alleviate disease in the 5xFAD mouse model of Alzheimer's disease. The Journal of Neuroscience, 43(37), 6460-6475.
Scopus42 WoS40 Europe PMC372023 Gartner, M. J., Lee, L. Y. Y., Mordant, F. L., Suryadinata, R., Chen, J., Robinson, P., . . . Subbarao, K. (2023). Ancestral, Delta, and Omicron (BA.1) SARS-CoV-2 strains are dependent on serine proteases for entry throughout the human respiratory tract. Med, 4(12), 944-955.e7.
Scopus8 WoS9 Europe PMC92023 Perlin, J. R., Anderson, W. J., Bartfeld, S., Couturier, A., de Soysa, Y., Hawley, R. S., . . . Mazzoni, E. O. (2023). ISSCR Education Committee syllabus and learning guide for enhancing stem cell literacy. Stem Cell Reports, 18(2), 417-419.
Scopus2 WoS2 Europe PMC32023 Tuano, N. K., Beesley, J., Manning, M., Shi, W., Perlaza-Jimenez, L., Malaver-Ortega, L. F., . . . Rosenbluh, J. (2023). CRISPR screens identify gene targets at breast cancer risk loci. Genome Biology, 24(1), 59.
Scopus15 WoS14 Europe PMC212023 Heazlewood, S. Y., Ahmad, T., Cao, B., Cao, H., Domingues, M., Sun, X., . . . Nilsson, S. K. (2023). High ploidy large cytoplasmic megakaryocytes are hematopoietic stem cells regulators and essential for platelet production. Nature communications, 14(1), 18 pages.
Scopus22 WoS21 Europe PMC222023 Foong, D., Mikhael, M., Zhou, J., Zarrouk, A., Liu, X., Schröder, J., . . . O’Connor, M. D. (2023). Transcriptome and Proteome Profiling of Primary Human Gastric Interstitial Cells of Cajal Predicts Pacemaker Networks. Journal of Neurogastroenterology and Motility, 29(2), 238-249.
Scopus2 WoS2 Europe PMC12023 Hawdon, A., Geoghegan, N. D., Mohenska, M., Elsenhans, A., Ferguson, C., Polo, J. M., . . . Zenker, J. (2023). Apicobasal RNA asymmetries regulate cell fate in the early mouse embryo. Nature Communications, 14(1), 1-18.
Scopus12 WoS12 Europe PMC162023 Leinenga, G., Bodea, L. G., Schröder, J., Sun, G., Zhou, Y., Song, J., . . . Götz, J. (2023). Transcriptional signature in microglia isolated from an Alzheimer's disease mouse model treated with scanning ultrasound. Bioengineering and Translational Medicine, 8(1), e10329-1-e10329-14.
Scopus13 WoS13 Europe PMC142022 Nefzger, C. M., Jardé, T., Srivastava, A., Schroeder, J., Rossello, F. J., Horvay, K., . . . Polo, J. M. (2022). Intestinal stem cell aging signature reveals a reprogramming strategy to enhance regenerative potential. NPJ Regenerative medicine, 7(1), 31-1-31-8.
Scopus9 WoS9 Europe PMC92022 Chothani, S. P., Adami, E., Widjaja, A. A., Langley, S. R., Viswanathan, S., Pua, C. J., . . . Schafer, S. (2022). A high-resolution map of human RNA translation. Molecular Cell, 82(15), 2885-2899.
Scopus77 WoS73 Europe PMC942022 Vallejo, A., Harvey, K., Wang, T., Wise, K., Butler, L., Polo, J., . . . Martelotto, L. (2022). snPATHO-seq: unlocking the FFPE archives for single nucleus RNA profiling.
Europe PMC92022 Huyghe, A., Furlan, G., Schroeder, J., Cascales, E., Trajkova, A., Ruel, M., . . . Lavial, F. (2022). Comparative roadmaps of reprogramming and oncogenic transformation identify Bcl11b and Atoh8 as broad regulators of cellular plasticity. Nature cell biology, 24(9), 1350-1363.
Scopus19 WoS20 Europe PMC212022 Choo, X. Y., McInnes, L. E., Grubman, A., Wasielewska, J. M., Belaya, I., Burrows, E., . . . White, A. R. (2022). Novel Anti-Neuroinflammatory Properties of a Thiosemicarbazone-Pyridylhydrazone Copper(II) Complex. International Journal of Molecular Sciences, 23(18), 1-27.
Scopus14 WoS14 Europe PMC72022 Tan, J. P., Liu, X., & Polo, J. M. (2022). Establishment of human induced trophoblast stem cells via reprogramming of fibroblasts. Nature Protocols, 17(12), 2739-2759.
Scopus20 WoS19 Europe PMC172022 Herring, C. A., Simmons, R. K., Freytag, S., Poppe, D., Moffet, J. J. D., Pflueger, J., . . . Lister, R. (2022). Human prefrontal cortex gene regulatory dynamics from gestation to adulthood at single-cell resolution. Cell, 185(23), 4428-4447.e28.
Scopus95 WoS91 Europe PMC1332022 Gerdes, P., Lim, S. M., Ewing, A. D., Larcombe, M. R., Chan, D., Sanchez-Luque, F. J., . . . Faulkner, G. J. (2022). Retrotransposon instability dominates the acquired mutation landscape of mouse induced pluripotent stem cells.. Nat Commun, 13(1), 18 pages.
Scopus12 WoS11 Europe PMC152022 Chang, Y. -C., Manent, J., Schroeder, J., Wong, S. F. L., Hauswirth, G. M., Shylo, N. A., . . . McGlinn, E. (2022). Nr6a1 controls Hox expression dynamics and is a master regulator of vertebrate trunk development. Nature Communications, 13(1), 1-19.
Scopus17 WoS16 Europe PMC172022 Lao, J. C., Bui, C. B., Pang, M. A., Cho, S. X., Rudloff, I., Elgass, K., . . . Nold, M. F. (2022). Type 2 immune polarization is associated with cardiopulmonary disease in preterm infants. Science Translational Medicine, 14(639), eaaz8454-1-eaaz8454-17.
Scopus25 WoS24 Europe PMC242022 Keniry, A., Jansz, N., Gearing, L. J., Wanigasuriya, I., Chen, J., Nefzger, C. M., . . . Blewitt, M. E. (2022). BAF complex-mediated chromatin relaxation is required for establishment of X chromosome inactivation. Nature Communications, 13(1), 1-15.
Scopus7 WoS6 Europe PMC122022 Larcombe, M. R., Hsu, S., Polo, J. M., & Knaupp, A. S. (2022). Indirect Mechanisms of Transcription Factor-Mediated Gene Regulation during Cell Fate Changes.. Advanced genetics (Hoboken, N.J.), 3(4), 2200015.
Europe PMC32022 Hauswirth, G. M., Garside, V. C., Wong, L. S. F., Bildsoe, H., Manent, J., Chang, Y. -C., . . . McGlinn, E. (2022). Breaking constraint of mammalian axial formulae. Nature Communications, 13(1), 1-12.
Scopus13 WoS12 Europe PMC82022 Mohenska, M., Tan, N. M., Tokolyi, A., Furtado, M. B., Costa, M. W., Perry, A. J., . . . Ramialison, M. (2022). 3D-cardiomics: A spatial transcriptional atlas of the mammalian heart. Journal of Molecular and Cellular Cardiology, 163, 20-32.
Scopus14 WoS13 Europe PMC182021 Nicholls, J., Cao, B., Le Texier, L., Xiong, L. Y., Hunter, C. R., Llanes, G., . . . MacDonald, K. P. A. (2021). Bone marrow regulatory T cells are a unique population, supported by niche-specific cytokines and plasmacytoid dendritic cells, and required for chronic graft-versus-host disease control. Frontiers in Cell and Developmental Biology, 9, 737880-1-737880-20.
Scopus13 WoS12 Europe PMC102021 Wang, J., Farkas, C., Benyoucef, A., Carmichael, C., Haigh, K., Wong, N., . . . Haigh, J. J. (2021). Interplay between the EMT transcription factors ZEB1 and ZEB2 regulates hematopoietic stem and progenitor cell differentiation and hematopoietic lineage fidelity. PLoS Biology, 19(9), e3001394-1-e3001394-33.
Scopus27 WoS25 Europe PMC272021 Fatima, S., Wagstaff, K. M., Lim, S. M., Polo, J. M., Young, J. C., & Jans, D. A. (2021). The nuclear transporter importin 13 is critical for cell survival during embryonic stem cell differentiation. Biochemical and Biophysical Research Communications, 534, 141-148.
Scopus5 WoS4 Europe PMC42021 Grubman, A., Choo, X. Y., Chew, G., Ouyang, J. F., Sun, G., Croft, N. P., . . . Polo, J. M. (2021). Transcriptional signature in microglia associated with Aβ plaque phagocytosis. Nature Communications, 12(1), 3015-1-3015-22.
Scopus195 WoS192 Europe PMC2192021 Liu, X., Tan, J. P., Schröder, J., Aberkane, A., Ouyang, J. F., Mohenska, M., . . . Polo, J. M. (2021). Modelling human blastocysts by reprogramming fibroblasts into iBlastoids. Nature, 591(7851), 627-632.
Scopus276 WoS267 Europe PMC2312021 Sun, X., Cao, B., Naval-Sanchez, M., Pham, T., Sun, Y. B. Y., Williams, B., . . . Nilsson, S. K. (2021). Nicotinamide riboside attenuates age-associated metabolic and functional changes in hematopoietic stem cells. Nature Communications, 12(1), 1-17.
Scopus83 WoS96 Europe PMC692021 Williamson, A., Liyanage, S., Hassanshahi, M., Dona, M., Toledo-Flores, D., Tran, D. X. A., . . . Psaltis, P. (2021). Discovery of embryonically derived bipotent endothelial-macrophage progenitor cells in postnatal aorta.
2020 Liu, X., Ouyang, J. F., Rossello, F. J., Tan, J. P., Davidson, K. C., Valdes, D. S., . . . Polo, J. M. (2020). Reprogramming roadmap reveals route to human induced trophoblast stem cells. Nature, 586(7827), 101-[107].
Scopus152 WoS142 Europe PMC1512020 Lee, K. M., Hawi, Z. H., Parkington, H. C., Parish, C. L., Kumar, P. V., Polo, J. M., . . . Tong, J. (2020). The application of human pluripotent stem cells to model the neuronal and glial components of neurodevelopmental disorders. Molecular Psychiatry, 25(2), 368-378.
Scopus32 WoS28 Europe PMC272020 Knaupp, A. S., Mohenska, M., Larcombe, M. R., Ford, E., Lim, S. M., Wong, K., . . . Polo, J. M. (2020). TINC — A Method to Dissect Regulatory Complexes at Single-Locus Resolution — Reveals an Extensive Protein Complex at the Nanog Promoter. Stem Cell Reports, 15(6), 1246-1259.
Scopus11 WoS10 Europe PMC112020 Kamaraj, U. S., Chen, J., Katwadi, K., Ouyang, J. F., Yang Sun, Y. B., Lim, Y. M., . . . Rackham, O. J. L. (2020). EpiMogrify models H3K4me3 data to identify signaling molecules that improve cell fate control and maintenance. Cell Systems, 11(5), 509-522.
Scopus13 WoS12 Europe PMC82020 Jardé, T., Chan, W. H., Rossello, F. J., Kaur Kahlon, T., Theocharous, M., Kurian Arackal, T., . . . Abud, H. E. (2020). Mesenchymal niche-derived neuregulin-1 drives intestinal stem cell proliferation and regeneration of damaged epithelium. Cell Stem Cell, 27(4), 646-662.e7.
Scopus107 WoS102 Europe PMC1152020 Chew, S., Lampinen, R., Saveleva, L., Korhonen, P., Mikhailov, N., Grubman, A., . . . Kanninen, K. M. (2020). Urban air particulate matter induces mitochondrial dysfunction in human olfactory mucosal cells. Particle and Fibre Toxicology, 17(1), 1-15.
Scopus44 WoS40 Europe PMC312020 Grubman, A., Vandekolk, T. H., Schröder, J., Sun, G., Hatwell-Humble, J., Chan, J., . . . Polo, J. M. (2020). A CX3CR1 reporter hESC line facilitates integrative analysis of in-vitro-derived microglia and improved microglia identity upon neuron-glia co-culture. Stem Cell Reports, 14(6), 1018-1032.
Scopus20 WoS21 Europe PMC222020 Oikari, L. E., Pandit, R., Stewart, R., Cuní-López, C., Quek, H., Sutharsan, R., . . . White, A. R. (2020). Altered brain endothelial cell phenotype from a familial Alzheimer mutation and its potential implications for amyloid clearance and drug delivery. Stem Cell Reports, 14(5), 924-939.
Scopus62 WoS60 Europe PMC602020 Covello, G., Rossello, F. J., Filosi, M., Gajardo, F., Duchemin, A. L., Tremonti, B. F., . . . Poggi, L. (2020). Transcriptome analysis of the zebrafish atoh7−/− Mutant, lakritz, highlights Atoh7-dependent genetic networks with potential implications for human eye diseases. FASEB BioAdvances, 2(7), 434-448.
Scopus5 WoS5 Europe PMC62019 Seruggia, D., Oti, M., Tripathi, P., Canver, M. C., LeBlanc, L., Di Giammartino, D. C., . . . Das, P. P. (2019). TAF5L and TAF6L Maintain Self-Renewal of Embryonic Stem Cells via the MYC Regulatory Network. Molecular Cell, 74(6), 1148-1163.
Scopus35 WoS33 Europe PMC372019 Ouyang, J. F., Kamaraj, U. S., Polo, J. M., Gough, J., & Rackham, O. J. L. (2019). Erratum: Correction to: Molecular Interaction Networks to Select Factors for Cell Conversion (Methods in molecular biology (Clifton, N.J.) (2019) 1975 (333-361)). Methods in molecular biology (Clifton, N.J.), 1975, C1.
2019 Skvortsova, K., Masle-Farquhar, E., Luu, P. L., Song, J. Z., Qu, W., Zotenko, E., . . . Clark, S. J. (2019). DNA hypermethylation encroachment at CpG island borders in cancer is predisposed by H3K4 monomethylation patterns. Cancer Cell, 35(2), 297-314.e8.
Scopus65 WoS64 Europe PMC552019 Tong, J., Lee, K. M., Liu, X., Nefzger, C. M., Vijayakumar, P., Hawi, Z., . . . Bellgrove, M. A. (2019). Generation of four iPSC lines from peripheral blood mononuclear cells (PBMCs) of an attention deficit hyperactivity disorder (ADHD) individual and a healthy sibling in an Australia-Caucasian family. Stem Cell Research, 34, 101353.
Scopus11 WoS8 Europe PMC92019 Rackham, O. J. L., & Polo, J. M. (2019). Let it RE:IN: integrating experimental observations to predict pluripotency network behaviour. EMBO Journal, 38(1), 3 pages.
2019 Grubman, A., Chew, G., Ouyang, J. F., Sun, G., Choo, X. Y., McLean, C., . . . Polo, J. M. (2019). A single-cell atlas of entorhinal cortex from individuals with Alzheimer’s disease reveals cell-type-specific gene expression regulation. Nature Neuroscience, 22(12), 2087-2097.
Scopus632 WoS615 Europe PMC6792019 Roco, J. A., Mesin, L., Binder, S. C., Nefzger, C., Gonzalez-Figueroa, P., Canete, P. F., . . . Vinuesa, C. G. (2019). Class-switch recombination occurs infrequently in germinal centers. Immunity, 51(2), 337-350.e7.
Scopus376 WoS363 Europe PMC3442018 Pastor, W. A., Liu, W., Chen, D., Ho, J., Kim, R., Hunt, T. J., . . . Clark, A. T. (2018). TFAP2C regulates transcription in human naive pluripotency by opening enhancers. Nature Cell Biology, 20(5), 553-564.
Scopus131 WoS129 Europe PMC1462018 Chen, J., Nefzger, C. M., Rossello, F. J., Sun, Y. B. Y., Lim, S. M., Liu, X., . . . Barberi, T. (2018). Fine tuning of canonical wnt stimulation enhances differentiation of pluripotent stem cells independent of β-catenin-mediated T-cell factor signaling. Stem Cells, 36(6), 822-833.
Scopus13 WoS11 Europe PMC112018 Ratnadiwakara, M., Archer, S. K., Dent, C. I., De Los Mozos, I. R., Beilharz, T. H., Knaupp, A. S., . . . Anko, M. L. (2018). SRSF3 promotes pluripotency through Nanog mRNA export and coordination of the pluripotency gene expression program. eLife, 7, e37419-1-e37419-28.
Scopus45 WoS44 Europe PMC372018 Pflueger, C., Tan, D., Swain, T., Nguyen, T., Pflueger, J., Nefzger, C., . . . Lister, R. (2018). A modular dCas9-SunTag DNMT3A epigenome editing system overcomes pervasive off-target activity of direct fusion dCas9-DNMT3A constructs. Genome Research, 28(8), 1193-1206.
Scopus142 WoS130 Europe PMC1302018 La, H. M., Mäkelä, J. A., Chan, A. L., Rossello, F. J., Nefzger, C. M., Legrand, J. M. D., . . . Hobbs, R. M. (2018). Identification of dynamic undifferentiated cell states within the male germline. Nature Communications, 9(1), 1-18.
Scopus77 WoS76 Europe PMC672017 Knaupp, A. S., Buckberry, S., Pflueger, J., Lim, S. M., Ford, E., Larcombe, M. R., . . . Polo, J. M. (2017). Transient and Permanent Reconfiguration of Chromatin and Transcription Factor Occupancy Drive Reprogramming. Cell Stem Cell, 21(6), 834-845.
Scopus95 WoS91 Europe PMC952017 Nguyen, P. D., Gurevich, D. B., Sonntag, C., Hersey, L., Alaei, S., Nim, H. T., . . . Currie, P. D. (2017). Muscle stem cells undergo extensive clonal drift during tissue growth via meox1-mediated induction of G2 cell-cycle arrest. Cell Stem Cell, 21(1), 107-119.
Scopus66 WoS61 Europe PMC612017 Nefzger, C. M., Rossello, F. J., Chen, J., Liu, X., Knaupp, A. S., Firas, J., . . . Polo, J. M. (2017). Cell type of origin dictates the route to pluripotency. Cell Reports, 21(10), 2649-2660.
Scopus44 WoS39 Europe PMC402017 O’Brien, C. M., Chy, H. S., Zhou, Q., Blumenfeld, S., Lambshead, J. W., Liu, X., . . . Laslett, A. L. (2017). New monoclonal antibodies to defined cell surface proteins on human pluripotent stem cells. Stem Cells, 35(3), 626-640.
Scopus17 WoS15 Europe PMC162017 Mariño, E., McLeod, K., Stanley, D., Yap, Y., Knight, J., McKenzie, C., . . . Mackay, C. (2017). Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nature Immunology, 18(5), 552-562.
Scopus587 WoS539 Europe PMC4852017 Firas, J., & Polo, J. M. (2017). Towards understanding transcriptional networks in cellular reprogramming. Current Opinion in Genetics and Development, 46, 1-8.
Scopus3 WoS3 Europe PMC22017 Nefzger, C. M., & Polo, J. M. (2017). DEAD-Box RNA Binding Protein DDX5: Not a Black-Box during Reprogramming. Cell Stem Cell, 20(4), 419-420.
Scopus5 WoS5 Europe PMC42017 Liu, X., Nefzger, C. M., Rossello, F. J., Chen, J., Knaupp, A. S., Firas, J., . . . Polo, J. M. (2017). Comprehensive characterization of distinct states of human naive pluripotency generated by reprogramming. Nature Methods, 14(11), 1055-1062.
Scopus127 WoS128 Europe PMC1202016 Kamaraj, U. S., Gough, J., Polo, J. M., Petretto, E., & Rackham, O. J. L. (2016). Computational methods for direct cell conversion. Cell Cycle, 15(24), 3343-3354.
Scopus10 WoS10 Europe PMC102016 Alaei, S., Knaupp, A. S., Lim, S. M., Chen, J., Holmes, M. L., Änkö, M. L., . . . Polo, J. M. (2016). An improved reprogrammable mouse model harbouring the reverse tetracycline-controlled transcriptional transactivator 3. Stem Cell Research, 17(1), 49-53.
Scopus9 WoS8 Europe PMC92016 Nefzger, C. M., Jardé, T., Rossello, F. J., Horvay, K., Knaupp, A. S., Powell, D. R., . . . Polo, J. M. (2016). A versatile strategy for isolating a highly enriched population of intestinal stem cells. Stem Cell Reports, 6(3), 321-329.
Scopus26 WoS25 Europe PMC222016 Rackham, O. J. L., Firas, J., Fang, H., Oates, M. E., Holmes, M. L., Knaupp, A. S., . . . Gough, J. (2016). A predictive computational framework for direct reprogramming between human cell types. NATURE GENETICS, 48(3), 331-335.
Scopus217 WoS196 Europe PMC1982015 Horvay, K., Jardé, T., Casagranda, F., Perreau, V. M., Haigh, K., Nefzger, C. M., . . . Abud, H. E. (2015). Snai1 regulates cell lineage allocation and stem cell maintenance in the mouse intestinal epithelium. EMBO Journal, 34(10), 1319-1335.
Scopus48 WoS45 Europe PMC442015 Firas, J., Liu, X., Lim, S. M., & Polo, J. M. (2015). Transcription factor-mediated reprogramming: Epigenetics and therapeutic potential. Immunology and Cell Biology, 93(3), 284-289.
Scopus19 WoS18 Europe PMC162015 Shu, R., Wong, W., Ma, Q. H., Yang, Z. Z., Zhu, H., Liu, F. J., . . . Xiao, Z. C. (2015). APP intracellular domain acts as a transcriptional regulator of miR-663 suppressing neuronal differentiation. Cell Death and Disease, 6(2), 1-12.
Scopus57 WoS55 Europe PMC452014 Firas, J., Liu, X., & Polo, J. M. (2014). Epigenetic memory in somatic cell nuclear transfer and induced pluripotency: evidence and implications. Differentiation, 88(1), 29-32.
Scopus15 WoS14 Europe PMC132014 Firas, J., Liu, X., Nefzger, C. M., & Polo, J. M. (2014). GM-CSF and MEF-conditioned media support feeder-free reprogramming of mouse granulocytes to iPS cells. Differentiation, 87(5), 193-199.
Scopus10 WoS9 Europe PMC82014 Nefzger, C. M., Alaei, S., Knaupp, A. S., Holmes, M. L., & Polo, J. M. (2014). Cell surface marker mediated purification of iPS cell intermediates from a reprogrammable mouse model. Journal of Visualized Experiments, 91(91), 1-8.
Scopus17 WoS17 Europe PMC112014 David, L., & Polo, J. M. (2014). Phases of reprogramming. Stem Cell Research, 12(3), 754-761.
Scopus105 WoS98 Europe PMC892014 Hobbs, R. M., & Polo, J. M. (2014). Reprogramming can be a transforming experience. Cell Stem Cell, 14(3), 269-271.
Scopus5 WoS5 Europe PMC42014 Nguyen, P. D., Hollway, G. E., Sonntag, C., Miles, L. B., Hall, T. E., Berger, S., . . . Currie, P. D. (2014). Haematopoietic stem cell induction by somite-derived endothelial cells controlled by meox1. Nature, 512(7514), 314-318.
Scopus115 WoS116 Europe PMC1152013 Hatzi, K., Jiang, Y., Huang, C., Garrett-Bakelman, F., Gearhart, M. D., Giannopoulou, E. G., . . . Melnick, A. (2013). A hybrid mechanism of action for BCL6 in B cells defined by formation of functionally distinct complexes at enhancers and promoters. Cell Reports, 4(3), 578-588.
Scopus160 WoS154 Europe PMC1492013 Apostolou, E., Ferrari, F., Walsh, R. M., Bar-Nur, O., Stadtfeld, M., Cheloufi, S., . . . Hochedlinger, K. (2013). Genome-wide chromatin interactions of the nanog locus in pluripotency, differentiation, and reprogramming. Cell Stem Cell, 12(6), 699-712.
Scopus174 WoS165 Europe PMC1552013 Kaur, G., Costa, M. W., Nefzger, C. M., Silva, J., Fierro-González, J. C., Polo, J. M., . . . Plachta, N. (2013). Probing transcription factor diffusion dynamics in the living mammalian embryo with photoactivatable fluorescence correlation spectroscopy. Nature Communications, 4(1), 1-13.
Scopus74 WoS72 Europe PMC672013 De, S., Shaknovich, R., Riester, M., Elemento, O., Geng, H., Kormaksson, M., . . . Michor, F. (2013). Aberration in DNA Methylation in B-Cell Lymphomas Has a Complex Origin and Increases with Disease Severity. Plos Genetics, 9(1), 14 pages.
Scopus108 WoS101 Europe PMC922013 Kelly, R. D. W., Rodda, A. E., Dickinson, A., Mahmud, A., Nefzger, C. M., Lee, W., . . . St John, J. C. (2013). Mitochondrial DNA haplotypes define gene expression patterns in pluripotent and differentiating embryonic stem cells. Stem Cells, 31(4), 703-716.
Scopus66 WoS66 Europe PMC532012 Hansson, J., Rafiee, M. R., Reiland, S., Polo, J. M., Gehring, J., Okawa, S., . . . Krijgsveld, J. (2012). Highly Coordinated Proteome Dynamics during Reprogramming of Somatic Cells to Pluripotency. Cell Reports, 2(6), 1579-1592.
Scopus202 WoS195 Europe PMC1962012 Polo, J. M., Anderssen, E., Walsh, R. M., Schwarz, B. A., Nefzger, C. M., Lim, S. M., . . . Hochedlinger, K. (2012). A molecular roadmap of reprogramming somatic cells into iPS cells. Cell, 151(7), 1617-1632.
Scopus713 WoS682 Europe PMC6352011 Ohi, Y., Qin, H., Hong, C., Blouin, L., Polo, J. M., Guo, T., . . . Ramalho-Santos, M. (2011). Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nature Cell Biology, 13(5), 541-549.
Scopus500 WoS473 Europe PMC4402011 Arnold, K., Sarkar, A., Yram, M. A., Polo, J. M., Bronson, R., Sengupta, S., . . . Hochedlinger, K. (2011). Sox2 + adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell, 9(4), 317-329.
Scopus634 WoS601 Europe PMC5992010 Cerchietti, L. C., Ghetu, A. F., Zhu, X., Da Silva, G. F., Zhong, S., Matthews, M., . . . Melnick, A. (2010). A Small-Molecule Inhibitor of BCL6 Kills DLBCL Cells In Vitro and In Vivo. Cancer Cell, 17(4), 400-411.
Scopus263 WoS240 Europe PMC2252010 Buecker, C., Chen, H. H., Polo, J. M., Daheron, L., Bu, L., Barakat, T. S., . . . Geijsen, N. (2010). A murine ESC-like state facilitates transgenesis and homologous recombination in human pluripotent stem cells. Cell Stem Cell, 6(6), 535-546.
Scopus186 WoS173 Europe PMC1602010 Polo, J. M., & Hochedlinger, K. (2010). When fibroblasts MET iPSCs. Cell Stem Cell, 7(1), 5-6.
Scopus52 WoS48 Europe PMC512010 Hammerich-Hille, S., Kaipparettu, B. A., Tsimelzon, A., Creighton, C. J., Jiang, S., Polo, J. M., . . . Oesterreich, S. (2010). SAFB1 mediates repression of immune regulators and apoptotic genes in breast cancer cells. Journal of Biological Chemistry, 285(6), 3608-3616.
Scopus32 WoS28 Europe PMC312010 Polo, J. M., Liu, S., Figueroa, M. E., Kulalert, W., Eminli, S., Tan, K. Y., . . . Hochedlinger, K. (2010). Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nature Biotechnology, 28(8), 848-855.
Scopus1027 WoS925 Europe PMC8602010 Duy, C., Yu, J. J., Nahar, R., Swaminathan, S., Kweon, S. M., Polo, J. M., . . . Müschen, M. (2010). BCL6 is critical for the development of a diverse primary B cell repertoire. Journal of Experimental Medicine, 207(6), 1209-1221.
Scopus106 WoS101 Europe PMC982009 Ci, W., Polo, J. M., Cerchietti, L., Shaknovich, R., Wang, L., Shao, N. Y., . . . Melnick, A. (2009). The BCL6 transcriptional program features repression of multiple oncogenes in primary B cells and is deregulated in DLBCL. Blood, 113(22), 5536-5548.
Scopus198 WoS180 Europe PMC1762009 Juszczynski, P., Chen, L., O'Donnell, E., Polo, J. M., Ranuncolo, S. M., Dalla-Favera, R., . . . Shipp, M. A. (2009). BCL6 modulates tonic BCR signaling in diffuse large B-cell lymphomas by repressing the SYK phosphatase, PTPROt. Blood, 114(26), 5315-5321.
Scopus55 WoS53 Europe PMC512009 Cerchietti, L. C., Yang, S. N., Shaknovich, R., Hatzi, K., Polo, J. M., Chadburn, A., . . . Melnick, A. (2009). A peptomimetic inhibitor of BCL6 with potent antilymphoma effects in vitro and in vivo. Blood, 113(15), 3397-3405.
Scopus158 WoS142 Europe PMC1342009 Utikal, J., Polo, J. M., Stadtfeld, M., Maherali, N., Kulalert, W., Walsh, R. M., . . . Hochedlinger, K. (2009). Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature, 460(7259), 1145-1148.
Scopus735 WoS678 Europe PMC6062008 Ranuncolo, S. M., Wang, L., Polo, J. M., Dell'Oso, T., Dierov, J., Gaymes, T. J., . . . Melnick, A. (2008). BCL6-mediated attenuation of DNA damage sensing triggers growth arrest and senescence through a p53-dependent pathway in a cell context-dependent manner. Journal of Biological Chemistry, 283(33), 22565-22572.
Scopus39 WoS34 Europe PMC362008 Ci, W., Polo, J. M., & Melnick, A. (2008). B-cell lymphoma 6 and the molecular pathogenesis of diffuse large B-cell lymphoma. Current Opinion in Hematology, 15(4), 381-390.
Scopus72 WoS65 Europe PMC612008 Ranuncolo, S. M., Polo, J. M., & Melnick, A. (2008). BCL6 represses CHEK1 and suppresses DNA damage pathways in normal and malignant B-cells. Blood Cells Molecules and Diseases, 41(1), 95-99.
Scopus92 WoS78 Europe PMC692008 Cerchietti, L. C., Polo, J. M., Da Silva, G. F., Farinha, P., Shaknovich, R., Gascoyne, R. D., . . . Melnick, A. (2008). Sequential transcription factor targeting for diffuse large B-cell lymphomas. Cancer Research, 68(9), 3361-3369.
Scopus27 WoS25 Europe PMC282008 Mendez, L. M., Polo, J. M., Yu, J. J., Krupski, M., Ding, B. B., Melnick, A., & Ye, B. H. (2008). CtBP is an essential corepressor for BCL6 autoregulation. Molecular and Cellular Biology, 28(7), 2175-2186.
Scopus58 WoS54 Europe PMC562008 Polo, J. M., Ci, W., Licht, J. D., & Melnick, A. (2008). Reversible disruption of BCL6 repression complexes by CD40 signaling in normal and malignant B cells. Blood, 112(3), 644-651.
Scopus38 WoS35 Europe PMC352007 Polo, J. M., Juszczynski, P., Monti, S., Cerchietti, L., Ye, K., Greally, J. M., . . . Melnick, A. (2007). Transcriptional signature with differential expression of BCL6 target genes accurately identifies BCL6-dependent diffuse large B cell lymphomas. Proceedings of the National Academy of Sciences of the United States of America, 104(9), 3207-3212.
Scopus122 WoS112 Europe PMC1002007 Parekh, S., Polo, J. M., Shaknovich, R., Juszczynski, P., Lev, P., Ranuncolo, S. M., . . . Melnick, A. (2007). BCL6 programs lymphoma cells for survival and differentiation through distinct biochemical mechanisms. Blood, 110(6), 2067-2074.
Scopus123 WoS114 Europe PMC1022007 Ranuncolo, S. M., Polo, J. M., Dierov, J., Singer, M., Kuo, T., Greally, J., . . . Melnick, A. (2007). Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nature Immunology, 8(7), 705-714.
Scopus238 WoS215 Europe PMC2122004 Polo, J. M., Dell'Oso, T., Ranuncolo, S. M., Cerchietti, L., Beck, D., Da Silva, G. F., . . . Melnick, A. (2004). Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nature Medicine, 10(12), 1329-1335.
Scopus261 WoS248 Europe PMC2202004 Evelson, P., Llesuy, S., Filinger, E., Rodriguez, R. R., Lemberg, A., Scorticati, C., . . . Perazzo, J. C. (2004). DECREASED OXIDATIVE STRESS IN PREHEPATIC PORTAL HYPERTENSIVE RAT LIVERS FOLLOWING THE INDUCTION OF DIABETES. Clinical and Experimental Pharmacology and Physiology, 31(3), 169-173.
2001 Travacio, M., Marı́a Polo, J., & Llesuy, S. (2001). Erratum to “Chromium (VI) induces oxidative stress in the mouse brain”. Toxicology, 162(2), 139-148.
2000 Ansaldo, M. X. N., Luquet, C. M., Evelson, P. A., Polo, J. X. M., & Llesuy, S. (2000). Antioxidant levels from different Antarctic fish caught around South Georgia Island and Shag Rocks. Polar Biology, 23(3), 160-165.
-
Book Chapters
Year Citation 2023 Tan, J. P., Liu, X., & Polo, J. M. (2023). In vitro models of human blastocysts and early embryogenesis. In P. C. K. Leung, & J. Qiao (Eds.), Human Reproductive and Prenatal Genetics (2 ed., pp. 311-328). Elsevier.
DOI2022 Onfray, C., Tan, J. P., Kilens, S., Liu, X., Polo, J., & David, L. (2022). Induction of Human Naïve Pluripotent Stem Cells from Somatic Cells. In P. Rugg-Gunn (Ed.), Methods in Molecular Biology (Vol. 2416, pp. 39-51). Springer US.
DOI Scopus2 Europe PMC22022 Knaupp, A. S., Schittenhelm, R. B., & Polo, J. M. (2022). Characterization of Mammalian Regulatory Complexes at Single-Locus Resolution Using TINC. In J. Horsfield, & J. Marsman (Eds.), Chromatin: Methods and Protocols (Vol. 2458, pp. 175-193). New York, NY: Humana.
DOI Europe PMC12019 Ouyang, J. F., Kamaraj, U. S., Polo, J. M., Gough, J., & Rackham, O. J. L. (2019). Molecular interaction networks to select factors for cell conversion. In Methods in Molecular Biology (Vol. 1975, pp. 333-361). Springer New York.
DOI Scopus3 Europe PMC32019 Liu, X., Chen, J., Firas, J., Paynter, J. M., Nefzger, C. M., & Polo, J. M. (2019). Generation of mouse-induced pluripotent stem cells by lentiviral transduction. In Methods in Molecular Biology (Vol. 1940, pp. 63-76). Springer New York.
DOI Scopus3 Europe PMC32015 Nefzger, C. M., Alaei, S., & Polo, J. M. (2015). Isolation of reprogramming intermediates during generation of induced pluripotent stem cells from mouse embryonic fibroblasts. In Methods in Molecular Biology (Vol. 1330, pp. 205-218). Springer New York.
DOI Scopus3 Europe PMC3 -
Preprint
Year Citation 2024 Buckberry, S., Liu, X., Poppe, D., Tan, J. P., Faulkner, G., Polo, J., & Lister, R. (2024). Transient Naive Treatment (TNT) iPS cells do not feature Sendai virus expression: Response to Sendai virus persistence questions the transient naive reprogramming method for iPSC generation.
DOI2023 Ly, H., Chung, J., Nguyen, J. H. V., Tian, L., Schroeder, J., Knaupp, A., . . . Ryall, J. (2023). Metabolism regulates muscle stem cell self-renewal by connecting the microenvironment and histone acetylation.
DOI Europe PMC12023 Yap, K., Schröder, J., Gerrand, Y. -W., Kong, A., Fox, A., Knowles, B., . . . Mitchell, G. (2023). Liver-specification of human iPSC-derived endothelial cells transplanted into mouse liver.
DOI2023 Chowdhury, M. M., Zimmerman, S., Leeson, H., Nefzger, C. M., Mar, J. C., Laslett, A., . . . Cooper-White, J. J. (2023). Substrate stiffness facilitates improved induced pluripotent stem cell production through modulation of both early and late phases of cell reprogramming.
DOI2023 Healy, E., Zhang, Q., Gail, E., Agius, S., Sun, G., Bullen, M., . . . Davidovich, C. (2023). The apparent loss of PRC2 chromatin occupancy as an artefact of RNA depletion.
DOI Europe PMC12022 Namipashaki, A., Pugsley, K., Liu, X., Abrehart, K., Lim, S. M., Sun, G., . . . Hawi, Z. (2022). Integration of Xeno-Free Single-cell Cloning in CRISPR-mediated DNA Editing of Human iPSCs Improves Homogeneity and Methodological Efficiency of Cellular Disease Modelling.
DOI2022 Chang, Y. -C., Lisa Wong, S. F., Schroeder, J., Hauswirth, G., Shylo, N., Moore, E., . . . McGlinn, E. (2022). <i>Nr6a1</i> controls axially-restricted body elongation, segmentation, patterning and lineage allocation.
DOI2022 Gerdes, P., Lim, S. M., Ewing, A., Larcombe, M., Chan, D., Sanchez-Luque, F., . . . Faulkner, G. (2022). Retrotransposon instability dominates the acquired mutation landscape of mouse induced pluripotent stem cells.
DOI2022 Rosenbluh, J., Tuano, N., Beesley, J., Manning, M., Shi, W., Malaver-Ortega, L., . . . Chenevix-Trench, G. (2022). CRISPR screens identify gene targets and drug repositioning opportunities at breast cancer risk loci.
DOI2021 Chen, J., Neil, J. A., Tan, J. P., Rudraraju, R., Mohenska, M., Sun, Y. B. Y., . . . Polo, J. M. (2021). An iTSC-derived placental model of SARS-CoV-2 infection reveals ACE2-dependent susceptibility in syncytiotrophoblasts.
DOI Europe PMC22021 Tuano, N., Beesley, J., Manning, M., Shi, W., Malaver-Ortega, L., Paynter, J., . . . Rosenbluh, J. (2021). CRISPR screens identify gene targets and drug repositioning opportunities at breast cancer risk loci.
DOI2020 Huyghe, A., Furlan, G., Schroeder, J., Stüder, J., Mugnier, F., De Matteo, L., . . . Lavial, F. (2020). The comprehensive roadmaps of reprogramming and transformation unveiled antagonistic roles for bHLH transcription factors in the control of cellular plasticity.
DOI2020 Covello, G., Rossello, F., Filosi, M., Gajardo, F., Duchemin, A. -L., Tremonti, B., . . . Poggi, L. (2020). Transcriptome analysis of the zebrafish<i>atoh7−/−</i>mutant,<i>lakritz</i>, highlights Atoh7-dependent genetic networks with potential implications for human eye diseases.
DOI2020 Knaupp, A. S., Mohenska, M., Larcombe, M. R., Ford, E., Lim, S. M., Wong, K., . . . Polo, J. M. (2020). TINC - a method to dissect transcriptional complexes at single locus resolution - reveals novel<i>Nanog</i>regulators in mouse embryonic stem cells.
DOI2019 Mohenska, M., Tan, N., Tokolyi, A., Furtado, M., Costa, M., Perry, A., . . . Polo, J. (2019). 3D-Cardiomics: A spatial transcriptional atlas of the mammalian heart.
DOI2019 Keniry, A., Jansz, N., Gearing, L., Wanigasuriya, I., Chen, J., Nefzger, C., . . . Blewitt, M. (2019). Xmas ESC: A new female embryonic stem cell system that reveals the BAF complex as a key regulator of the establishment of X chromosome inactivation.
DOI2019 Grubman, A., Choo, X. Y., Chew, G., Ouyang, J., Sun, G., Croft, N., . . . Polo, J. (2019). Mouse and human microglial phenotypes in Alzheimer’s disease are controlled by amyloid plaque phagocytosis through Hif1α.
DOI2019 Grubman, A., Chew, G., Ouyang, J., Sun, G., Choo, X. Y., McLean, C., . . . Polo, J. (2019). A single cell brain atlas in human Alzheimer’s disease.
DOI Europe PMC12018 Tong, J., Lee, K. M., Liu, X., Nefzger, C., Vijayakumar, P., Hawi, Z., . . . Bellgrove, M. (2018). Generation of four iPSC lines from peripheral blood mononuclear cells (PBMCs) of an Attention Deficit Hyperactivity Disorder (ADHD) individual and a healthy sibling in an Australia-Caucasian family.
DOI2018 Pflueger, C., Tan, D., Swain, T., Nguyen, T., Pflueger, J., Nefzger, C., . . . Lister, R. (2018). A modular dCas9-SunTag DNMT3A epigenome editing system overcomes pervasive off-target activity of direct fusion dCas9-DNMT3A constructs.
DOI
Research Grants
| Date | Details | Title |
|---|---|---|
| 2021–2023 | NHMRC, Ideas Grant, CIA | Developing an in vitro model of a human blastocyst |
| 2021–2023 | NHMRC, Ideas Grant, CIA | Reprogramming human fibroblasts into induced trophoblast stem cells |
| 2021-2023 | ARC, Discovery Project, CIA | How do transcription factors control cell fate transitions? |
| 2020–2021 | MRFF-Coronavirus Research Response–Rapid Screening of Approved Drugs in Stem Cell Models for COVID-19 Treatment, CI | Stem cell-derived human tissue models for the identification of drugs to treat COVID-19. |
| 2020–2023 | Department of Health and Human Services Victoria, DHHS | Evaluating direct and indirect effects of SARS-CoV-2 on multiple organ systems using stem cell-derived human tissues |
| 2020-2022 | Cancer Council Victoria, PCI | Targeted reprogramming of Prostate Cancer |
| 2019–2020 | The CASS Foundation, CI | Targeting prostate cancer with maths. |
| 2018–2022 | ARC Future Fellowship |
Unveiling the epigenome dynamics through the pluripotency continuum The Fellowship supports Professor Polo salary. |
| 2018–2021 | NHMRC, CIB | Exploring and Targeting the Anti-Inflammatory Signaling Mechanisms of IL-3. |
| 2018–2020 | ARC Discovery Project, CIB | Regulatory architecture of the trunk-to-tail transition. |
| 2017-2020 | NHMRC, CIB | Leveraging genomics strategies to generate adult neurons from iPSCs and somatic cells. |
| 2017–2018 | Prostate Cancer Foundation of Australia, CI | A predictive computational framework for targeted reprogramming of castrate resistant prostate cancer. |
| 2016–2019 | NHMRC, CIA | Unveiling the human reprogramming pathway. |
| 2016–2019 | NHMRC, CIA | Using direct reprogramming to generate and rejuvenate Haematopoietic Stem Cells |
| 2015-2019 | NHMRC, CIB | Bone Marrow Endothelial Stem Cells have the capacity to form both the endothelial and haematopoietic hierarchies. |
| 2015-2017 | NHMRC, CIA | Inducing and controlling cellular plasticity. |
| 2014-2016 | NHMRC, CIA | Epigenetic and functional decline of intestinal stem cells during aging. |
| 2014–2018 | Sylvia & Charles Viertel Foundation Senior Medical Research Fellowship | The Fellowship supported Professor Polo salary from 2014-1018 |
| 2014-2016 | NHMRC, CIB | Advancing regenerative medicine by epigenetic engineering of human induced pluripotent stem cells. |
| 2014-2018 | Stem Cells Australia, Principal Investigator | |
| 2014 | ARC LIEF, Principal Investigator | This Grant underpinned the establishment of a Single Cell Genomic Centre |
| 2013-2015 | NHMRC, CIA | Determining how the germ layer of origin of adult somatic cells influences the differentiation potential of induced pluripotent stem cells. |
| 2013-2015 | NHMRC, CIA | Does nuclear reprogramming of granulocytes induce reversal of the haematopoiesis pathway. |
| 2012-2015 | NHMRC, Chief Investigator, CDF | The Fellowship supported Professor Polo salary from 2012-2013, it was discontinued upon being granted the Sylvia & Charles Viertel Foundation Senior Medical Research Fellowship. |
Teaching, Mentoring and Research Supervision
| Postdoctoral Fellows (Present) |
|---|
| Dr. Sandii Constable (Lab Manager) |
| Dr. Naiara Bediaga |
| Dr. German Moran |
|
|
| Postdoctoral Fellows (Past) |
|---|
| Dr. Christian Nefzger (Now group leader at UQ) |
| Dr. Fernando Rossello (Now Level C at University of Melbourne) |
| Dr. Kathryn Davidson (Now Project Manager at WEHI) |
| Dr. Alexandra Grubman (Now Medical Scientific Liaison at Biogen) |
| Dr. Jan Manent (Now postdoc at ARMI) |
| Higher Degree Research Graduate students |
|---|
| Monika Mohenska (Faculty Medicine, Nursing and Health Sciences PhD Program): Developing an algorithm to predict cellular identities given a spatial niche. |
| Joseph Chen (Industrial Partnership Fellowship) Using TFs to enhance induced pluripotency and direct reprogramming. |
| Jacob Michael Paynter (Faculty Medicine, Nursing and Health Sciences PhD Program): Generating human haematopoietic stem cells via transcription factor mediated reprogramming. |
| Michael Larcombe (Faculty Medicine, Nursing and Health Sciences PhD Program): Dissecting the regulatory complexes controlling pluripotency factors. |
| Jia Ping Tan (Faculty Medicine, Nursing and Health Sciences PhD Program): Understanding the molecular control and cell fate transitions of human induced trophoblast stem cells. |
| Esther Louise Miriklis (Australian Regenerative Medicine Institute): Spatial gene regulation via single cell analyses and advanced fluorescence microscopy. |
| Postgraduate students trained to date (PhD) | Year |
|---|---|
| Xiaodong Ethan Liu (Monash Postgraduate Award Scholarship): Integrative molecular analyses reveal distinct reprogramming trajectories into states of naïve and primed human induced pluripotency. He was awarded the top 100 thesis from a Chinese student abroad by the Chinese government in 2019. Awarded the Stem Cell Biology Ridolfo award for best student of the year in 2018. Due to COVID19, he is doing a short postdoc in the Polo Lab. | 2019 |
| Xin Yi Choo (International Postgraduate Research Scholarship): Development of novel therapeutic approaches for treatment of Alzheimer’s disease. Now postdoc at Duke-NUS, Singapore. | 2018 |
| Jaber Firas: Towards an understanding of induced cellular plasticity. He was awarded the VC thesis Commendation. Awarded the Stem Cell Biology Ridolfo award for best student of the year in 2016. Now training as MD to become a Physician-Scientist. | 2018 |
| Suzan DeBoer: Modelling epigenetic dysregulation in developmental disorders. Now a postdoc in Leiden University Medical Center, Holland. | 2017 |
| Sara Alaei: Dynamics of transcription factor targeting and chromatin states during reprogramming. Now Postdoc at ARMI | 2016 |
| Bianca Borchin: Directed derivation and FACS-mediated purification of PAX3+/PAX7+ skeletal muscle precursors from human pluripotent stem cells. Professor Polo supervised her last year and thesis writing due to her first supervisor leaving the Institute. Bianca is now a Postdoc in Memorial Sloan Kettering. | 2015 |
| Honours students trained to date | Year |
|---|---|
| Kayla Wong | 2018 |
| Jia Tan | 2018 |
| Matt Tiedemann | 2017 |
| Amber Rucinski | 2016 |
| Nathalia Tan | 2015 |
| Mitchel de Souza | 2014 |
| Jaber Firas | 2013 |
| Masters Student | Year |
|---|---|
| Daniela Pufleau Valdes (Master of Biomedical & Health Science): Molecular and functional characterisation of iTSCs derived from reprogrammed human fibroblasts. | 2019 - 2020 |
| International Masters student | Year |
|---|---|
| Frida Forsberg (Uppsala University) | 2015 |
| Research in Action undergraduate student | Year |
|---|---|
| Neiloy Chowdhury | 2018 |
| Raymond Zhang | 2018 |
| Sheree Chen | 2018 |
| Pranjal Patel | 2018 |
| Natalie Saunders | 2017 |
| Kayla Wong | 2017 |
| Kesh Faye-Chauhan | 2013 |
| Laura Corfield | 2013 |
| Erica Min Joung Kim | 2012 |
| Research Assistant |
|---|
| Dr. Guizhi Sun |
| Research Assistants (Past) |
|---|
| Dr. Melissa Holmes |
| Ketan Mishra |
| Margeaux Hodgson-Garms |
| Joseph Chen |
| Jacob Paynter |
| Michael Larcombe |
-
Current Higher Degree by Research Supervision (University of Adelaide)
Date Role Research Topic Program Degree Type Student Load Student Name 2025 Co-Supervisor Integrating multi-omics data to identify novel biomarkers for improved disease diagnosis and prognosis Doctor of Philosophy Doctorate Full Time Miss Anxuan Han 2024 Co-Supervisor The role of coding and non-coding RNA in atherosclerotic plaque phenotype in diabetic vs non-diabetic patients with peripheral arterial disease Doctor of Philosophy Doctorate Part Time Dr Benjamin Thurston 2023 Principal Supervisor Somatic cell reprogramming of fibroblasts from different mammals to model early development. Doctor of Philosophy Doctorate Full Time Miss Elly Deidre Walters 2023 Principal Supervisor How pluripotent transcription factors reshape the prostate cancer epigenetic landscape to reprogram the cell fate toward neuroendocrine-like with their cofactors Doctor of Philosophy Doctorate Full Time Mr Tianjun Zhang
-
Committee Memberships
Date Role Committee Institution Country 2020 - ongoing Chair ARMI Group Leader Meeting ARMI Australia 2019 - ongoing Chair Academic Steering Committee Monash Micromon Genomics Platform Australia 2019 - ongoing Member Co-organiser of the Single Cell Symposium, DUKE-NUS, Singapore Singapore 2019 - ongoing Member International Society for Stem Cell Research (ISSCR) Education Committee International Society for Stem Cell Research (ISSCR) Australia 2019 - ongoing Member Monash Proteomics Platform Steering Committee Monash Proteomics Australia 2017 - ongoing Co-Chair Reprogramming Symposium Stem Cells Stream at ComBio - ASBMB 2017 Australia 2017 - 2019 Member Pluripotency Stream Leader and member of the Scientific Leadership Group at ARC Stem Cells Australia Special Initiative. Australia 2016 - 2018 Member Department of Anatomy and Developmental Biology Executive Committee Department of Anatomy and Developmental Biology Australia 2016 - ongoing Chair Chair and Programme Co-organizer, Margaret River Regional Forum/ASSCR, 2017 Margaret River Regional Forum/ASSCR Australia 2015 - 2018 Member ARMI HDR committee ARMI Australia 2014 - ongoing Chair Lead organizer of the Australasian Society for Stem Cell Research Annual Meeting. Australasian Society for Stem Cell Research Australia 2014 - ongoing Chair Pluripotency Symposium, Stem Cells Stream at ComBio - ASBMB 2014 Stem Cells Stream Australia 2014 - 2017 Member Faculty of Biomedical and Psychological Sciences (FBPS) Gender Equity Committee Faculty of Biomedical and Psychological Sciences (FBPS) Australia 2013 - 2018 Member MHTP Bioinformatics Committee Monash Institute of Medical Research Australia 2013 - 2017 Member Cell Reprogramming Australia Cell Reprogramming Australia Australia 2013 - 2015 Co-Chair Cell Reprogramming Australia Annual Conference Cell Reprogramming Australia Australia -
Editorial Boards
Date Role Editorial Board Name Institution Country 2020 - ongoing Member Stem Cell Reports (Official Journal of the International Society for Stem Cell Research) International Society for Stem Cell Research Australia -
Review, Assessment, Editorial and Advice
Date Title Type Institution Country 2016 - ongoing Silvia and Charles Viertel Foundation (External) Peer Review Silvia and Charles Viertel Foundation (External) - 2016 - ongoing Biotechnology and Biological Sciences Research Council (UK) Peer Review Biotechnology and Biological Sciences Research Council (UK) - 2013 - ongoing Fondation pour la Recherche Médicale (France) Peer Review Fondation pour la Recherche Médicale (France) - 2013 - ongoing Reviewer for Academic Promotion Peer Review University of Melbourne, University of Edinburgh, Tokyo University, MRC LMS, Imperial College, John Hopkins University - 2012 - ongoing Neurological Foundation of New Zealand Peer Review Neurological Foundation of New Zealand - 2011 - ongoing NHMRC (Panel member and external) and ARC. Peer Review NHMRC and ARC -
Connect With Me
External Profiles





