Dr David Bersten
Grant-Funded Researcher (B)
School of Pharmacy and Biomedical Sciences
College of Health
Eligible to supervise Masters and PhD - email supervisor to discuss availability.
bHLH-PAS transcription factors are critical biological sensors of physiological (hypoxia, tryptophan metabolites, neuronal activity, and appetite) and environmental (diet derived metabolites and environmental pollutants) stimuli to homeostatically regulate genes involved in adaptation. My research focuses on an achieving and understanding of how transcription and gene regulation controls normal and pathological function, with a particular focus on the central nervous system. Using a combination of molecular techniques, mouse and cellular models I study the control of gene regulation in response to hypoxia, neuronal activity and appetite. Defining Target Gene Selection RulesbHLH-PAS transcription factors execute discrete biological functions through regulation of distinct target genes. The mechanisms by which bHLH-PAS transcription factors select different target genes is largely unknown. Through integrated genomic and proteomic analysis we aim to define the rules by which transcription factors select and activate highly specific target gene expression. We are investigating the contribution of inherent DNA binding activities (both in vitro and in vivo) and the role of chromatin and DNA associated proteins and modifications in regulating site occupancy and selecting transcription factor specific target genes. The results of this work will provide a structural frame work to define the target gene selection rules for mammalian transcription factors. Transcription and gene regulation in central nervous system development and function.Neuronal PAS transcription factors control brain excitabilityThe Neuronal PAS transcription factors (NPAS1, NPAS3, NPAS4) collectively regulate the neuron network activity balance (excitatory and inhibitory inputs) in the brain, and, as such are implicated in diseases where activity imbalance is observed (Autism, Schizophrenia, and Seizures). NPAS4 mRNA and protein is rapidly and temporally regulated by neuronal activity (depolarisation) in both glutamatergic and GABAergic neurons to homeostatically regulate synaptic plasticity and memory formation. While NPAS1and NPAS3 have apposing roles in generation of GABAergic interneurons and inhibitory neuron signalling. We aim to understand the regulation of the expression and activity of the neuronal PAS transcription factors to regulate neuron network activity, providing key mechanistic insights into how dysfunction in neuronal activity can lead to disease. Hypothalamic transcription factors control appetite disorders in humansThe hypothalamus is responsible for the regulation of many goal orientate behaviours including energy homeostasis, hunger and satiety signalling. It contains small numbers highly specialised neuropeptide expressing neurons that relay peripheral circulating signals to the brain eliciting a homeostatic change in animal behaviour. We have found that two hypothalamically restricted transcription factors Single minded 1 (Sim1), a bHLH-PAS transcription factor and Orthopedia (OTP) a homeodomain transcription factor are mutated in individuals with severe obesity and Prader-Willi-like features (Sim1). We have also shown that knock-in mouse models containing these mutations phenocopy the severe obesity seen in human patients. We are therefore interested in the understanding the how transcription factor mediated changes in gene expression lead to ravenous hunger. GABA A receptor epsilon and its intronic encoded microRNA cluster miR-224/452 control neurotransmitter specification and function.GABA is the main inhibitory neuron transmitter in the central nervous system and primarily acts through a pentameric GABA A receptor made up of 2 alpha subunits, 2 beta subunits and a gamma subunit to form a GABA responsive Cl- permeable ion channel. The GABRE subunit (a gamma-like subunit) has been shown to have a number of unique properties including conferring an insensitivity of the GABA A receptor to anaesthetics (pregnanolone and pentobarbital). We have found that GABRE is expressed in AGRP and POMC neuropeptide expressing neurons of the hypothalamus and hindbrain serotonergic neurons. We are currently investigating the role of GABRE and miR-224/452 in the development and function of neuronal subtypes in the brain.
bHLH-PAS transcription factors are critical biological sensors of physiological (hypoxia, tryptophan metabolites, neuronal activity, and appetite) and environmental (diet derived metabolites and environmental pollutants) stimuli to homeostatically regulate genes involved in adaptation. My research focuses on an achieving and understanding of how transcription and gene regulation controls normal and pathological function, with a particular focus on the central nervous system. Using a combination of molecular techniques, mouse and cellular models I study the control of gene regulation in response to hypoxia, neuronal activity and appetite.
Defining Target Gene Selection Rules
bHLH-PAS transcription factors execute discrete biological functions through regulation of distinct target genes. The mechanisms by which bHLH-PAS transcription factors select different target genes is largely unknown. Through integrated genomic and proteomic analysis we aim to define the rules by which transcription factors select and activate highly specific target gene expression. We are investigating the contribution of inherent DNA binding activities (both in vitro and in vivo) and the role of chromatin and DNA associated proteins and modifications in regulating site occupancy and selecting transcription factor specific target genes. The results of this work will provide a structural frame work to define the target gene selection rules for mammalian transcription factors.
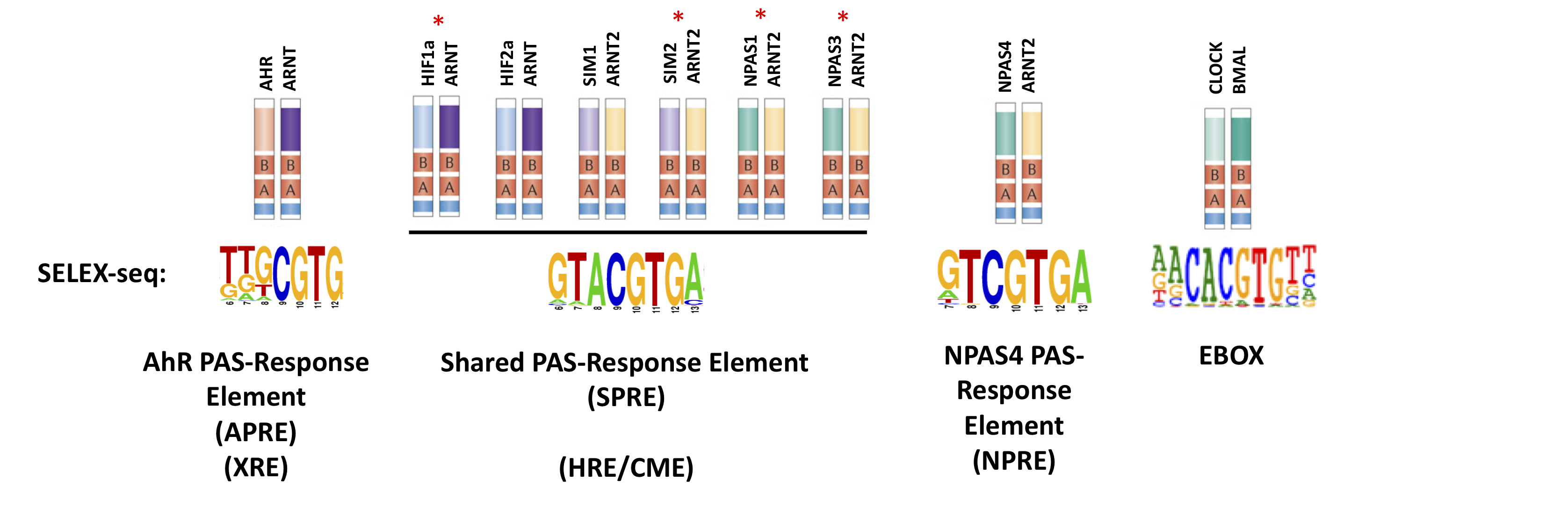
Transcription and gene regulation in central nervous system development and function.
Neuronal PAS transcription factors control brain excitability
The Neuronal PAS transcription factors (NPAS1, NPAS3, NPAS4) collectively regulate the neuron network activity balance (excitatory and inhibitory inputs) in the brain, and, as such are implicated in diseases where activity imbalance is observed (Autism, Schizophrenia, and Seizures). NPAS4 mRNA and protein is rapidly and temporally regulated by neuronal activity (depolarisation) in both glutamatergic and GABAergic neurons to homeostatically regulate synaptic plasticity and memory formation. While NPAS1and NPAS3 have apposing roles in generation of GABAergic interneurons and inhibitory neuron signalling. We aim to understand the regulation of the expression and activity of the neuronal PAS transcription factors to regulate neuron network activity, providing key mechanistic insights into how dysfunction in neuronal activity can lead to disease.
Hypothalamic transcription factors control appetite disorders in humans
The hypothalamus is responsible for the regulation of many goal orientate behaviours including energy homeostasis, hunger and satiety signalling. It contains small numbers of highly specialised neuropeptide expressing neurons that relay peripheral circulating signals to the brain eliciting a homeostatic change in animal behaviour. We have found that two hypothalamically restricted transcription factors Single minded 1 (Sim1), a bHLH-PAS transcription factor and Orthopedia (OTP) a homeodomain transcription factor are mutated in individuals with severe obesity and Prader-Willi-like features (Sim1). We have also shown that knock-in mouse models containing these mutations phenocopy the severe obesity seen in human patients. We are therefore interested in the understanding the how transcription factor mediated changes in gene expression lead to ravenous hunger.
GABA A receptor epsilon and its intronic encoded microRNA cluster miR-224/452 control neurotransmitter specification and function.
GABA is the main inhibitory neuron transmitter in the central nervous system and primarily acts through a pentameric GABA A receptor made up of 2 alpha subunits, 2 beta subunits and a gamma subunit to form a GABA responsive Cl- permeable ion channel. The GABRE subunit (a gamma-like subunit) has been shown to have a number of unique properties including conferring an insensitivity of the GABA A receptor to anaesthetics (pregnanolone and pentobarbital). We have found that GABRE is expressed in AGRP and POMC neuropeptide expressing neurons of the hypothalamus and hindbrain serotonergic neurons. We are currently investigating the role of GABRE and miR-224/452 in the development and function of neuronal subtypes in the brain.
| Date | Position | Institution name |
|---|---|---|
| 2017 - ongoing | Post Doctoral Research Fellow | The University of Adelaide |
| Date | Type | Title | Institution Name | Country | Amount |
|---|---|---|---|---|---|
| 2015 | Award | Dean’s Commendation for Doctoral Thesis Excellence | The University Of Adelaide | Australia | - |
| Date | Institution name | Country | Title |
|---|---|---|---|
| 2014 | The University of Adelaide | Australia | PhD Sciences |
| The University of Adelaide | Australia | BSc (Biomedical Science) | |
| The University of Adelaide | Australia | BSc (Honours) (Biochemistry) 1st Class |
| Year | Citation |
|---|---|
| - | Roennfeldt, A., Allen, T., Trowbridge, B., Beard, M., Whitelaw, M., Russell, D., . . . Peet, D. (n.d.). Graph Pad prism files of all data published in Roennfeldt et al. (2023). Data points for each experiment represents the mean of an individual replicate (and thus each experiment was completed in triplicate with the mean +/- standard deviation graphed).. DOI |
| Year | Citation |
|---|---|
| 2023 | Allen, T., Roennfeldt, A., Reckdharajkumar, M., Liu, M., Quinn, R., Russell, D., . . . Bersten, D. (2023). dFLASH; dual FLuorescent transcription factor Activity Sensor for Histone integrated live-cell reporting and high-content screening. DOI |
Identifying pathological mechanisms underlying intellectual disability (2015)
- Investigators: Jolly L; Gecz J; Whitelaw M; Bersten D
I currently lecture in the 3rd year Biochemistry course - Cancer, Stem Cells, and Development.
I lecture on neuronal development, directed differentiation of fibroblasts to functional neurons and molecular techniques to map, mark and test neuronal circuits.
| Date | Role | Research Topic | Program | Degree Type | Student Load | Student Name |
|---|---|---|---|---|---|---|
| 2022 | Co-Supervisor | Developing cell-based screening systems to identify small molecule inhibitors of the hypoxic signalling protein FIH, and to investigate the signalling pathways of ovulation. | Doctor of Philosophy | Doctorate | Full Time | Miss Alison Elizabeth Roennfeldt |
| 2022 | Co-Supervisor | Developing cell-based screening systems to identify small molecule inhibitors of the hypoxic signalling protein FIH, and to investigate the signalling pathways of ovulation. | Doctor of Philosophy | Doctorate | Full Time | Miss Alison Elizabeth Roennfeldt |
| Date | Role | Research Topic | Program | Degree Type | Student Load | Student Name |
|---|---|---|---|---|---|---|
| 2019 - 2024 | Co-Supervisor | Development and application of dual fluorescent reporting systems for HIF-1 alpha and other transcription factor pathways | Doctor of Philosophy | Doctorate | Full Time | Mr Timothy Patrick Allen |
| Date | Role | Research Topic | Location | Program | Supervision Type | Student Load | Student Name |
|---|---|---|---|---|---|---|---|
| 2018 - 2018 | Principal Supervisor | Cell based drug screening systems for the bHLH-PAS transcription factors | The Univeristy of Adelaidre | - | Honours | Full Time | Timothy Allen |
| 2017 - ongoing | Co-Supervisor | bHLH PAS and HD protein interactions in neuronal development and pathology | The Univeristy of Adelaide | - | Doctorate | Full Time | Joesph Rossi |
| 2016 - 2016 | Co-Supervisor | Characterising the transcriptional control of neuronal PAS domain factors NPAS1 and NPAS3 | The University Of Adelaide | - | Honours | Full Time | Victoria Nankivell |
| 2016 - 2016 | Co-Supervisor | Investigating Interactions Between the Neuronal Factors SIM1, OTP and MAGEL2 | The University Of Adelaide | - | Honours | - | Joseph Rossi |
| 2015 - 2016 | Co-Supervisor | Expression and Isolation of Mammalian Basic Helix-Loop-Helix Per-Arnt-Sim Domains in Escherichia coli | The University Of Adelaide | - | Master | Full Time | Chan Hoi Chong |
| 2014 - 2014 | Co-Supervisor | Expression of the SIM-ARNT protein complex using baculovirus | The University Of Adelaide | - | Master | Full Time | Muyang Wang |
| 2013 - 2013 | Co-Supervisor | bHLH-PAS Protein Knockdown Using Inducible and Reversible miR30-based shRNA Systems and in silico Model Generation for Potent shRNA Prediction | The University Of Adelaide | - | Master | Full Time | Dian Li |
| Date | Topic | Presented at | Institution | Country |
|---|---|---|---|---|
| 2018 - ongoing | Lecturing Biochemistry III - From Fibroblasts to Functional Neurons | - | The Univerity Of Adelaide | Australia |
| 2017 - ongoing | bHLH-PAS transcription factors: target gene selection, satiety and the homeodomain connection | COMBIO 2017 | The University Of Adelaide | Australia |
| 2012 - ongoing | REGULATION OF EXPRESSION AND ACTIVITY OF THE NEURONAL TRANSCRIPTION FACTOR NPAS4 | COMBIO 2012 | The University of Adelaide | - |




