Dr Anna Sheppard
Senior Lecturer
School of Biological Sciences
College of Sciences
Dr Anna Sheppard is a Lecturer in Bioinformatics at the University of Adelaide. Her research uses bacterial whole-genome sequencing to investigate the molecular epidemiology and transmission of bacterial pathogens both within outbreaks, as well as across geographic and ecological barriers within a One Health framework. She has a particular focus on antimicrobial resistance dissemination driven by mobile genetic elements, and she has developed novel bioinformatic methods for resistance gene tracking. Her research has been published in leading journals including the New England Journal of Medicine, Nature Microbiology, and the Proceedings of the National Academy of Sciences.Anna completed her PhD at the University of Adelaide in 2009, before undertaking postdoctoral training at Kiel University, Germany, and the University of Oxford, UK. She returned to the University of Adelaide as a Lecturer in the School of Biological Sciences in December 2021.
Modern medicine is heavily dependent on the availability of effective antibiotics. The increasing prevalence of antimicrobial resistance (AMR) in bacterial pathogens threatens to push medicine towards a ‘post-antibiotic’ era, where bacterial infections can no longer be reliably treated. Our research aims to improve understanding of bacterial genome evolution driving AMR dissemination, with the longer-term goal of using this information for designing interventions to reduce the spread of resistance.
Research in the Sheppard lab focuses particularly on ‘mobile AMR genes’ that can be transferred horizontally and confer resistance in different bacterial species. Mobility of these genes is driven by associations with different types of mobile genetic elements (MGEs), including:
- Plasmids, which drive horizontal transfer between different bacteria
- Transposons, which drive intracellular mobilisation from one genomic location to another (e.g. one plasmid to another, or from a plasmid to the chromosome)
- Integrons, which can capture multiple AMR genes, generating multi-drug resistance regions
AMR genes can be contained within multiple nested MGEs, creating ‘Russian Doll-like’ hierarchies, where each MGE is capable of independent mobilisation (Fig. 1). Over time, activity of different MGEs drives AMR gene spread across different plasmids and to different host bacteria. Because many different MGEs can be involved, AMR gene spread is associated with varied, and often complex, genomic rearrangements.
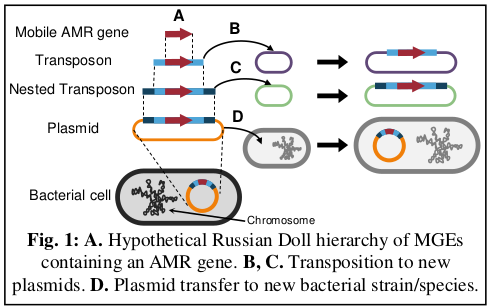
Our research uses whole-genome sequencing of bacterial pathogens to investigate mobile AMR genes and their associated MGEs. We use both short-read and long-read sequencing technologies. Long-read sequencing is particularly important for determining the genetic contexts of AMR genes because many MGEs are repetitive and therefore assemble poorly using short-read data. With long-read whole-genome sequencing we can routinely assemble complete, fully closed bacterial genomes, with a single contig for the bacterial chromosome and one contig for each plasmid.
We are particularly interested in determining dissemination pathways of mobile AMR genes. This is important both for characterising short-term transmission pathways for guiding infection control, as well as broader dissemination across geographic, ecological and species barriers for guiding strategies to tackle resistance within a One Health framework. In addition, we are interested in developing approaches for AMR gene tracking in the context of AMR surveillance.
Our research aims to:
- Determine how associations with different MGEs impact AMR spread
- Determine how MGE activity impacts bacterial genome evolution
- Improve biological understanding of MGEs driving AMR gene mobility
- Inform optimal sequence-based surveillance strategies for tracking critical AMR genes
- Develop software for AMR gene tracking using whole-genome sequencing data
Student Opportunities
Postgraduate and undergraduate student opportunities are available in the Sheppard lab, and we welcome students from both biological and computational backgrounds. While some previous bioinformatics experience is preferable, it is not necessary.
| Date | Position | Institution name |
|---|---|---|
| 2023 - ongoing | Senior Lecturer in Bioinformatics | University of Adelaide |
| 2021 - 2022 | Lecturer in Bioinformatics | University of Adelaide |
| 2020 - 2021 | Postdoctoral Researcher | University of Oxford |
| 2016 - 2018 | Visiting Assistant Professor | University of Virginia |
| 2013 - 2020 | Bioinformatician | University of Oxford |
| 2010 - 2012 | Postdoctoral Researcher | Kiel University |
| 2005 - 2006 | Research Officer | University of Adelaide |
| Date | Institution name | Country | Title |
|---|---|---|---|
| 2006 - 2009 | University of Adelaide | Australia | PhD |
| 2001 - 2004 | University of Adelaide | Australia | Bachelor of Science (Honours) |
| Year | Citation |
|---|---|
| 2026 | Cross, B. J., Partridge, S. R., & Sheppard, A. E. (2026). Impacts of mobile genetic elements on antimicrobial resistance genes in gram-negative pathogens: Current insights and genomic approaches. Microbiological Research, 302, 14 pages. Scopus4 WoS4 |
| 2025 | Yassine, I., Jolley, K. A., Bray, J. E., Jansen van Rensburg, M. J., Patel, F., Sheppard, A. E., . . . Brueggemann, A. B. (2025). Investigating the population structure of Moraxella catarrhalis using a cgMLST scheme and LIN code system.. Nat Commun, 16(1), 9137. |
| 2025 | Sunmonu, G. T., Lo, S. W., Sheppard, A. E., & Ogunniyi, A. D. (2025). Serotype replacement and mobile genetic elements in Streptococcus pneumoniae: a systematic review.. Microb Genom, 11(9), 17 pages. |
| 2024 | Matthews, C. A., Watson-Haigh, N. S., Burton, R. A., & Sheppard, A. E. (2024). A gentle introduction to pangenomics. Briefings in Bioinformatics, 25(6), 13 pages. Scopus15 WoS13 Europe PMC14 |
| 2024 | Bouras, G., Houtak, G., Wick, R. R., Mallawaarachchi, V., Roach, M. J., Papudeshi, B., . . . Vreugde, S. (2024). Hybracter: enabling scalable, automated, complete and accurate bacterial genome assemblies. Microbial Genomics, 10(5), 001244-1-001244-15. Scopus54 WoS52 Europe PMC58 |
| 2023 | Davies, T. J., Swann, J., Sheppard, A. E., Pickford, H., Lipworth, S., AbuOun, M., . . . Stoesser, N. (2023). Discordance between different bioinformatic methods for identifying resistance genes from short-read genomic data, with a focus on Escherichia coli. Microbial genomics, 9(12), 11 pages. Scopus5 WoS5 Europe PMC6 |
| 2023 | Bouras, G., Sheppard, A. E., Mallawaarachchi, V., & Vreugde, S. (2023). Plassembler: an automated bacterial plasmid assembly tool.. Bioinformatics (Oxford, England), 39(7), btad409. Scopus37 WoS44 Europe PMC51 |
| 2021 | Matlock, W., Chau, K. K., AbuOun, M., Stubberfield, E., Barker, L., Kavanagh, J., . . . Woodford, N. (2021). Genomic network analysis of environmental and livestock F-type plasmid populations. The ISME Journal, 15(8), 2322-2335. Scopus31 Europe PMC32 |
| 2021 | AbuOun, M., Jones, H., Stubberfield, E., Gilson, D., Shaw, L. P., Hubbard, A. T. M., . . . Anjum, M. F. (2021). A genomic epidemiological study shows that prevalence of antimicrobial resistance in Enterobacterales is associated with the livestock host, as well as antimicrobial usage. Microbial Genomics, 7(10). |
| 2021 | Shaw, L. P., Chau, K. K., Kavanagh, J., AbuOun, M., Stubberfield, E., Gweon, H. S., . . . Sheppard, A. (2021). Niche and local geography shape the pangenome of wastewater- and livestock-associated Enterobacteriaceae. Science Advances, 7(15). |
| 2020 | Mathers, A. J., Vegesana, K., German-Mesner, I., Ainsworth, J., Pannone, A., Crook, D. W., . . . Eyre, D. W. (2020). Risk factors for Klebsiella pneumoniae carbapenemase (KPC) gene acquisition and clinical outcomes across multiple bacterial species. Journal of Hospital Infection, 104(4), 456-468. Scopus30 Europe PMC24 |
| 2020 | Davies, T. J., Stoesser, N., Sheppard, A. E., Abuoun, M., Fowler, P., Swann, J., . . . Walker, A. S. (2020). Reconciling the Potentially Irreconcilable? Genotypic and Phenotypic Amoxicillin-Clavulanate Resistance in Escherichia coli. Antimicrobial Agents and Chemotherapy, 64(6), 1-16. Scopus38 Europe PMC39 |
| 2020 | David, S., Cohen, V., Reuter, S., Sheppard, A. E., Giani, T., Parkhill, J., . . . Aanensen, D. M. (2020). Integrated chromosomal and plasmid sequence analyses reveal diverse modes of carbapenemase gene spread among Klebsiella pneumoniae. Proceedings of the National Academy of Sciences, 117(40), 25043-25054. Scopus118 Europe PMC126 |
| 2019 | Gweon, H. S., Shaw, L. P., Swann, J., De Maio, N., AbuOun, M., Niehus, R., . . . Stoesser, N. (2019). The impact of sequencing depth on the inferred taxonomic composition and AMR gene content of metagenomic samples. Environmental Microbiome, 14(1). Scopus81 |
| 2019 | Elliott, Z. S., Barry, K. E., Cox, H. L., Stoesser, N., Carroll, J., Vegesana, K., . . . Mathers, A. J. (2019). The Role of fosA in Challenges with Fosfomycin Susceptibility Testing of Multispecies Klebsiella pneumoniae Carbapenemase-Producing Clinical Isolates. Journal of Clinical Microbiology, 57(10), 1-8. Scopus37 WoS36 Europe PMC29 |
| 2019 | De Maio, N., Shaw, L. P., Hubbard, A., George, S., Sanderson, N. D., Swann, J., . . . Woodford, N. (2019). Comparison of long-read sequencing technologies in the hybrid assembly of complex bacterial genomes. MICROBIAL GENOMICS, 5(9), 12 pages. Scopus191 WoS182 Europe PMC187 |
| 2019 | Rooney, C. M., Sheppard, A. E., Clark, E., Davies, K., Hubbard, A. T. M., Sebra, R., . . . Chilton, C. H. (2019). Dissemination of multiple carbapenem resistance genes in an in vitro gut model simulating the human colon. Journal of Antimicrobial Chemotherapy, 74(7), 1876-1883. Scopus26 WoS25 Europe PMC22 |
| 2019 | Mathers, A. J., Crook, D., Vaughan, A., Barry, K. E., Vegesana, K., Stoesser, N., . . . Sheppard, A. E. (2019). Klebsiella quasipneumoniae Provides a Window into Carbapenemase Gene Transfer, Plasmid Rearrangements, and Patient Interactions with the Hospital Environment. Antimicrobial Agents and Chemotherapy, 63(6), 1-12. Scopus66 WoS71 Europe PMC56 |
| 2019 | Barry, K. E., Wailan, A. M., Sheppard, A. E., Crook, D., Vegesana, K., Stoesser, N., . . . Mathers, A. J. (2019). Don't overlook the little guy: An evaluation of the frequency of small plasmids co-conjugating with larger carbapenemase gene containing plasmids. Plasmid, 103, 1-8. Scopus35 WoS35 Europe PMC36 |
| 2019 | van Aartsen, J. J., Moore, C. E., Parry, C. M., Turner, P., Phot, N., Mao, S., . . . Stoesser, N. (2019). Epidemiology of paediatric gastrointestinal colonisation by extended spectrum cephalosporin-resistant Escherichia coli and Klebsiella pneumoniae isolates in north-west Cambodia. BMC Microbiology, 19(1), 1-14. Scopus17 WoS17 Europe PMC18 |
| 2018 | Sheppard, A. E., Stoesser, N., German-Mesner, I., Vegesana, K., Walker, A. S., Crook, D. W., & Mathers, A. J. (2018). TETyper: a bioinformatic pipeline for classifying variation and genetic contexts of transposable elements from short-read whole-genome sequencing data. Microbial Genomics, 4(12), 1-9. Scopus34 WoS33 Europe PMC37 |
| 2018 | Eyre, D. W., Sheppard, A. E., Madder, H., Moir, I., Moroney, R., Quan, T. P., . . . Jeffery, K. J. M. (2018). A Candida auris Outbreak and Its Control in an Intensive Care Setting. NEW ENGLAND JOURNAL OF MEDICINE, 379(14), 1322-1331. Scopus421 WoS390 Europe PMC358 |
| 2018 | Barkham, T., Sheppard, A., Jones, N., & Chen, S. L. (2018). Streptococcus agalactiae that caused meningitis in healthy adults in 1998 are ST283, the same type that caused a food-borne outbreak of invasive sepsis in 2015: an observational molecular epidemiology study. Clinical Microbiology and Infection, 24(8), 923-925. Scopus6 WoS6 Europe PMC6 |
| 2018 | George, S., Pankhurst, L., Hubbard, A., Votintseva, A., Stoesser, N., Sheppard, A. E., . . . Phan, H. T. T. (2018). Resolving plasmid structures in Enterobacteriaceae using the MinION nanopore sequencer: assessment of MinION and MinION/Illumina hybrid data assembly approaches (vol 3, 2017). MICROBIAL GENOMICS, 4(3), 1 page. Scopus1 WoS1 |
| 2018 | Zhong, L. -L., Phan, H. T. T., Shen, C., Vihta, K. -D., Sheppard, A. E., Huang, X., . . . Tian, G. -B. (2018). High Rates of Human Fecal Carriage of mcr-1-Positive Multidrug-Resistant Enterobacteriaceae Emerge in China in Association With Successful Plasmid Families. CLINICAL INFECTIOUS DISEASES, 66(5), 676-685. Scopus63 WoS59 Europe PMC60 |
| 2017 | Hollensteiner, J., Poehlein, A., Sproeer, C., Bunk, B., Sheppard, A. E., Rosentstiel, P., . . . Liesegang, H. (2017). Complete Genome sequence of the nematicidal Bacillus thuringiensis MYBT18246. Standards in Genomic Sciences, 12(1), 1-10. Scopus8 WoS7 Europe PMC3 |
| 2017 | Hollensteiner, J., Poehlein, A., Sproeer, C., Bunk, B., Sheppard, A. E., Rosenstiel, P., . . . Liesegang, H. (2017). Complete genome sequence of the nematicidal Bacillus thuringiensis MYBT18247. JOURNAL OF BIOTECHNOLOGY, 260, 48-52. Scopus10 WoS10 Europe PMC5 |
| 2017 | Stoesser, N., Sheppard, A. E., Peirano, G., Anson, L. W., Pankhurst, L., Sebra, R., . . . Pitout, J. D. (2017). Genomic epidemiology of global Klebsiella pneumoniae carbapenemase (KPC)-producing Escherichia coli. SCIENTIFIC REPORTS, 7(1), 11 pages. Scopus94 WoS95 Europe PMC84 |
| 2017 | Orlek, A., Phan, H., Sheppard, A. E., Doumith, M., Ellington, M., Peto, T., . . . Stoesser, N. (2017). A curated dataset of complete Enterobacteriaceae plasmids compiled from the NCBI nucleotide database. Data in Brief, 12, 423-426. Scopus33 WoS33 Europe PMC36 |
| 2017 | Cheruvanky, A., Stoesser, N., Sheppard, A. E., Crook, D. W., Hoffman, P. S., Weddle, E., . . . Mathers, A. J. (2017). Enhanced Klebsiella pneumoniae Carbapenemase Expression from a Novel Tn4401 Deletion. ANTIMICROBIAL AGENTS AND CHEMOTHERAPY, 61(6), 10 pages. Scopus47 WoS48 Europe PMC49 |
| 2017 | Orlek, A., Phan, H., Sheppard, A. E., Doumith, M., Ellington, M., Peto, T., . . . Stoesser, N. (2017). Ordering the mob: Insights into replicon and MOB typing schemes from analysis of a curated dataset of publicly available plasmids. Plasmid, 91, 42-52. Scopus61 WoS58 Europe PMC60 |
| 2017 | Mathers, A. J., Stoesser, N., Chai, W., Carroll, J., Barry, K., Cherunvanky, A., . . . Sheppard, A. E. (2017). Chromosomal Integration of the Klebsiella pneumoniae Carbapenemase Gene, bla(KPC), in Klebsiella Species Is Elusive but Not Rare. Antimicrobial Agents and Chemotherapy, 61(3), 1-12. Scopus44 WoS44 Europe PMC43 |
| 2017 | Orlek, A., Stoesser, N., Anjum, M. F., Doumith, M., Ellington, M. J., Peto, T., . . . Sheppard, A. E. (2017). Plasmid Classification in an Era of Whole-Genome Sequencing: Application in Studies of Antibiotic Resistance Epidemiology. FRONTIERS IN MICROBIOLOGY, 8(FEB), 10 pages. Scopus159 WoS152 Europe PMC144 |
| 2017 | George, S., Pankhurst, L., Hubbard, A., Votintseva, A., Stoesser, N., Sheppard, A. E., . . . Phan, H. T. T. (2017). Resolving plasmid structures in Enterobacteriaceae using the MinION nanopore sequencer: assessment of MinION and MinION/Illumina hybrid data assembly approaches. MICROBIAL GENOMICS, 3(8), 8 pages. Scopus74 WoS70 Europe PMC66 |
| 2017 | Young, B. C., Wu, C. -H., Gordon, N. C., Cole, K., Price, J. R., Liu, E., . . . Wilson, D. J. (2017). Severe infections emerge from commensal bacteria by adaptive evolution. ELIFE, 6, 25 pages. Scopus79 WoS78 Europe PMC96 |
| 2016 | Seale, A. C., Koech, A. C., Sheppard, A. E., Barsosio, H. C., Langat, J., Anyango, E., . . . Berkley, J. A. (2016). Maternal colonization with Streptococcus agalactiae and associated stillbirth and neonatal disease in coastal Kenya. NATURE MICROBIOLOGY, 1(7), 10 pages. Scopus92 WoS89 Europe PMC89 |
| 2016 | Sheppard, A. E., Stoesser, N., Wilson, D. J., Sebra, R., Kasarskis, A., Anson, L. W., . . . Mathers, A. J. (2016). Nested Russian Doll-Like Genetic Mobility Drives Rapid Dissemination of the Carbapenem Resistance Gene bla(KPC). ANTIMICROBIAL AGENTS AND CHEMOTHERAPY, 60(6), 3767-3778. Scopus231 WoS226 Europe PMC254 |
| 2016 | Stoesser, N., Sheppard, A. E., Pankhurst, L., De Maio, N., Moore, C. E., Sebra, R., . . . Crook, D. W. (2016). Evolutionary History of the Global Emergence of the Escherichia coli Epidemic Clone ST131. MBIO, 7(2), 15 pages. Scopus274 WoS266 Europe PMC277 |
| 2016 | Sheppard, A. E., Vaughan, A., Jones, N., Turner, P., Turner, C., Efstratiou, A., . . . Seale, A. C. (2016). Capsular Typing Method for Streptococcus agalactiae Using Whole-Genome Sequence Data. JOURNAL OF CLINICAL MICROBIOLOGY, 54(5), 1388-1390. Scopus35 WoS29 Europe PMC34 |
| 2016 | Senn, L., Clerc, O., Zanetti, G., Basset, P., Prod'hom, G., Gordon, N. C., . . . Blanc, D. S. (2016). The Stealthy Superbug: the Role of Asymptomatic Enteric Carriage in Maintaining a Long-Term Hospital Outbreak of ST228 Methicillin-Resistant Staphylococcus aureus. MBIO, 7(1), 9 pages. Scopus75 WoS72 Europe PMC66 |
| 2016 | Stoesser, N., Sheppard, A. E., Peirano, G., Sebra, R., Lynch, T., Anson, L., . . . Pitout, J. D. (2016). Complete Sequencing of Plasmids Containing bla(OXA-163) and bla(OXA-48) in Escherichia coli Sequence Type 131. ANTIMICROBIAL AGENTS AND CHEMOTHERAPY, 60(11), 6948-6951. Scopus16 WoS17 Europe PMC14 |
| 2016 | Stoesser, N., Sheppard, A. E., Peirano, G., Sebra, R. P., Lynch, T., Anson, L. W., . . . Pitout, J. D. (2016). First Report of bla(IMP-14) on a Plasmid Harboring Multiple Drug Resistance Genes in Escherichia coli Sequence Type 131. Antimicrobial Agents and Chemotherapy, 60(8), 5068-5071. Scopus15 WoS14 Europe PMC13 |
| 2016 | Hardiman, C. A., Weingarten, R. A., Conlan, S., Khil, P., Dekker, J. P., Mathers, A. J., . . . Frank, K. M. (2016). Horizontal transfer of carbapenemase-encoding plasmids and comparison with hospital epidemiology data. Antimicrobial Agents and Chemotherapy, 60(8), 4910-4919. Scopus71 WoS69 Europe PMC61 |
| 2016 | Sheppard, A. E., Stoesser, N., Sebra, R., Kasarskis, A., Deikus, G., Anson, L., . . . Mathers, A. J. (2016). Complete Genome Sequence of KPC-Producing Klebsiella pneumoniae Strain CAV1193.. Genome announcements, 4(1), 2 pages. Scopus18 WoS15 Europe PMC20 |
| 2016 | Sheppard, A. E., Nakad, R., Saebelfeld, M., Masche, A. C., Dierking, K., & Schulenburg, H. (2016). High instability of a nematicidal Cry toxin plasmid in Bacillus thuringiensis. Journal of Invertebrate Pathology, 133, 34-40. Scopus6 WoS6 Europe PMC7 |
| 2015 | Masri, L., Branca, A., Sheppard, A. E., Papkou, A., Laehnemann, D., Guenther, P. S., . . . Schulenburg, H. (2015). Host–pathogen coevolution: The selective advantage of Bacillus thuringiensis virulence and its cry toxin genes. Plos Biology, 13(6), 1-30. Scopus69 WoS68 Europe PMC66 |
| 2015 | Mathers, A. J., Stoesser, N., Sheppard, A. E., Pankhurst, L., Giess, A., Yeh, A. J., . . . Sifri, C. D. (2015). Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae at a single institution: Insights into endemicity from Whole-genome sequencing. Antimicrobial Agents and Chemotherapy, 59(3), 1656-1663. Scopus120 WoS113 Europe PMC116 |
| 2015 | Stoesser, N., Sheppard, A. E., Moore, C. E., Golubchik, T., Parry, C. M., Nget, P., . . . Walker, A. S. (2015). Extensive within-host diversity in fecally carried extended-spectrum-beta-lactamase-producing Escherichia coli Isolates: Implications for transmission analyses. Journal of Clinical Microbiology, 53(7), 2122-2131. Scopus65 WoS63 Europe PMC69 |
| 2014 | Stoesser, N., Giess, A., Batty, E. M., Sheppard, A. E., Walker, A. S., Wilson, D. J., . . . Joshi, S. (2014). Genome sequencing of an extended series of NDM-producing Klebsiella pneumoniae isolates from neonatal infections in a Nepali hospital characterizes the extent of community- Versus hospital- associated transmission in an endemic setting. Antimicrobial Agents and Chemotherapy, 58(12), 7347-7357. Scopus110 WoS109 Europe PMC117 |
| 2014 | Stoesser, N., Sheppard, A. E., Shakya, M., Sthapit, B., Thorson, S., Giess, A., . . . Crook, D. W. (2014). Dynamics of MDR Enterobacter cloacae outbreaks in a neonatal unit in Nepal: Insights using wider sampling frames and next-generation sequencing. Journal of Antimicrobial Chemotherapy, 70(4), 1008-1015. Scopus52 WoS45 Europe PMC37 |
| 2013 | Sheppard, A. E., Poehlein, A., Rosenstiel, P., Liesegang, H., & Schulenburg, H. (2013). Complete genome sequence of Bacillus thuringiensis strain 407 Cry-. Genome Announcements, 1(1), 2 pages. Scopus41 WoS21 Europe PMC33 |
| 2011 | Sheppard, A., Madesis, P., Lloyd, A., Day, A., Ayliffe, M., & Timmis, J. (2011). Introducing an RNA editing requirement into a plastid-localised transgene reduces but does not eliminate functional gene transfer to the nucleus. Plant Molecular Biology, 76(3-5), 299-309. Scopus14 WoS14 Europe PMC12 |
| 2009 | Sheppard, A., & Timmis, J. (2009). Instability of Plastid DNA in the Nuclear Genome. PLoS Genetics, 5(1), 323. Scopus56 WoS53 Europe PMC51 |
| 2008 | Sheppard, A., Ayliffe, M., Blatch, L., Day, A., Delaney, S., Khairul-Fahmy, N., . . . Timmis, J. (2008). Transfer of plastid DNA to the nucleus is elevated during male gametogenesis in tobacco. Plant Physiology, 148(1), 328-336. Scopus54 WoS53 Europe PMC43 |
| - | Matlock, W., Lipworth, S., Chau, K. K., AbuOun, M., Barker, L., Kavanagh, J., . . . Stoesser, N. (2023). Enterobacterales plasmid sharing amongst human bloodstream infections, livestock, wastewater, and waterway niches in Oxfordshire, UK. eLife, 12. |
| Year | Citation |
|---|---|
| 2025 | Hassan, N. T., Van Treeck, B., Rodríguez-Vargas, A., Sheppard, A. E., Collins, K., & Adelson, D. L. (2025). Lineage-Specific Evolution, Structural Diversity, and Activity of R2 Retrotransposons in Animals.. DOI |
| 2025 | Yassine, I., Jolley, K. A., Bray, J. E., Jansen van Rensburg, M. J., Patel, F., Sheppard, A. E., . . . Brueggemann, A. B. (2025). Understanding the population structure of <i>Moraxella catarrhalis</i> using core genome multilocus sequence typing (cgMLST) and a life identification number (LIN) code classification system. DOI |
| 2023 | Bouras, G., Houtak, G., Wick, R., Mallawaarachchi, V., Roach, M., Papudeshi, B., . . . Vreugde, S. (2023). Hybracter: Enabling Scalable, Automated, Complete and Accurate Bacterial Genome Assemblies. DOI |
| 2017 | Young, B. C., Wu, C. -H., Claire Gordon, N., Cole, K., Price, J. R., Liu, E., . . . Wilson, D. J. (2017). Severe infections emerge from the microbiome by adaptive evolution. DOI |
| Date | Role | Research Topic | Program | Degree Type | Student Load | Student Name |
|---|---|---|---|---|---|---|
| 2025 | Principal Supervisor | Characterising insertion site preferences in mobile genetic elements | Master of Philosophy | Master | Full Time | Mr Oliver John Williams |
| 2025 | Principal Supervisor | Investigating Antimicrobial Resistance Plasmids in Biosolids-Amended Soil Using Multi-Omics Approaches | Master of Philosophy | Master | Part Time | Miss Deannon Louise Branch |
| 2025 | Co-Supervisor | Identification of pneumococcal serotype-independent antigenic regions and mobile genetic elements driving antimicrobial resistance gene mobility | Doctor of Philosophy | Doctorate | Full Time | Mr Gabriel Temitope Sunmonu |
| 2025 | Principal Supervisor | Investigating Antimicrobial Resistance Plasmids in Biosolids-Amended Soil Using Multi-Omics Approaches | Master of Philosophy | Master | Part Time | Miss Deannon Louise Branch |
| 2025 | Co-Supervisor | Identification of pneumococcal serotype-independent antigenic regions and mobile genetic elements driving antimicrobial resistance gene mobility | Doctor of Philosophy | Doctorate | Full Time | Mr Gabriel Temitope Sunmonu |
| 2025 | Principal Supervisor | Characterising insertion site preferences in mobile genetic elements | Master of Philosophy | Master | Full Time | Mr Oliver John Williams |
| 2024 | Principal Supervisor | Characterisation of Mobile Genetic Elements Driving the Global Dissemination of Key Antimicrobial Resistance Genes | Doctor of Philosophy | Doctorate | Full Time | Miss Bethany Jane Cross |
| 2024 | Co-Supervisor | Investigating the drivers of pneumococcal pathogenesis | Doctor of Philosophy | Doctorate | Full Time | Miss Kate Patricia Whyte |
| 2024 | Principal Supervisor | Establishing an MGE-based network approach for identifying AMR inheritance clusters | Doctor of Philosophy | Doctorate | Full Time | Mr Max Docherty-Kenny |
| 2024 | Principal Supervisor | Characterisation of Antimicrobial Resistance Genes and Mobile Genetic Elements in Animal Pathogens | Doctor of Philosophy | Doctorate | Full Time | Mr Guan Xue Lee |
| 2024 | Principal Supervisor | Establishing an MGE-based network approach for identifying AMR inheritance clusters | Doctor of Philosophy | Doctorate | Full Time | Mr Max Docherty-Kenny |
| 2024 | Principal Supervisor | Characterisation of Mobile Genetic Elements Driving the Global Dissemination of Key Antimicrobial Resistance Genes | Doctor of Philosophy | Doctorate | Full Time | Miss Bethany Jane Cross |
| 2024 | Principal Supervisor | Characterisation of Antimicrobial Resistance Genes and Mobile Genetic Elements in Animal Pathogens | Doctor of Philosophy | Doctorate | Full Time | Mr Guan Xue Lee |
| 2024 | Co-Supervisor | Investigating the drivers of pneumococcal pathogenesis | Doctor of Philosophy | Doctorate | Full Time | Miss Kate Patricia Whyte |
| 2023 | Co-Supervisor | Implement a scalable, automated workflow for transposon annotation as part of the Ruminant T2T genome sequencing consortium | Doctor of Philosophy | Doctorate | Full Time | Miss Luan Zhong |
| 2022 | Co-Supervisor | The Evolution, Distribution, and Activity of R2 Elements | Doctor of Philosophy | Doctorate | Full Time | Ms Nozhat Tabassum Hassan |
| 2022 | Co-Supervisor | Combining machine-learning and rational design for engineering heme enzyme biocatalysts and biosensors. | Doctor of Philosophy | Doctorate | Full Time | Miss Alecia Rachel Gee |
| 2022 | Co-Supervisor | Investigating pangenome graphs in plants | Doctor of Philosophy | Doctorate | Full Time | Miss Chelsea Anne Matthews |
| 2022 | Co-Supervisor | Investigating pangenome graphs in plants | Doctor of Philosophy | Doctorate | Full Time | Miss Chelsea Anne Matthews |
| 2022 | Co-Supervisor | Combining machine-learning and rational design for engineering heme enzyme biocatalysts and biosensors. | Doctor of Philosophy | Doctorate | Full Time | Miss Alecia Rachel Gee |
| 2022 | Co-Supervisor | The Evolution, Distribution, and Activity of R2 Elements | Doctor of Philosophy | Doctorate | Full Time | Ms Nozhat Tabassum Hassan |





