
Research Interests
Cancer Cell Biology Epigenetics Bioinformatics Cancer Biology and Clinical OncologyDr Luke Isbel
Externally-Funded Senior Research Fellow
School of Pharmacy and Biomedical Sciences
College of Health
Eligible to supervise Masters and PhD - email supervisor to discuss availability.
Dr Luke Isbel relocated to Adelaide in 2023 to set up his own lab at the South Australian immunoGENomics Cancer Institute (SAiGENCI) and Adelaide Centre for Epigenetics (ACE). Luke’s vision for his research group is to bridge the fundamental and translational aspects of gene regulation (epigenetics) using a cross-disciplinary, ‘systems’ approach. He will tackle critical gaps in our understanding of molecular events by integrating a variety of models and tools to shed light on this critical area of biology.PhD, Masters and Hons. Degree project available for creative and inspired individuals!!!Dr Luke Isbel completed his PhD (2016) in Melbourne with Professor Emma Whitelaw, followed by a phenomenal postdoc in Switzerland within the lab of Professor Dirk Schübeler at the Friedrich Miescher Institute for Biomedical Research. Throughout this, he worked primarily within cutting edge models of gene regulation, keenly interested in concepts like ‘specificity’ and ‘phenotypic buffering’. The strengths of Luke’s lab include delving into epigenetic processes in stem cells, specializing in functional genomics and bioinformatics techniques to study molecular mechanisms at fundamental levels.
How is specificity established in gene regulation and how can we target it?
Epigenetics refers to the forces within our cells that can enable genes to be expressed at the right time, place and developmental setting. Essentially, it ensure genes that are meant to be active stay active, while genes that are meant to be inactive remain inactive. This intricate process allows multiple tissue and cell types to exist in our bodies that nevertheless all arises from the same genetic material. Our goal is to unravel the molecular mechanisms of how such epigenetic forces drive cellular identity.
Our particular focus centres on transcription factors, which are proteins that ‘read out’ DNA sequence. They are critical for directing the complex gene expression patterns that are established during development. In fact, transcription factors are frequent drivers of cancer when mutated. However, understanding their function in normal or disease settings is challenging because our genomes are not naked - they are wrapped up by histone proteins to form chromatin. An overwhelming amount of evidence suggests that various chromatin states can regulate gene expression, potentially by working hand-in-hand with transcription factors, although the detailed molecular mechanisms remain largely enigmatic.
We are intrigued by interactions between transcription factors and chromatin. By targeting this interface, we aim to identify novel means of altering transcription factor activity to open new avenues of cancer treatment. To achieve this goal, we are using a variety of genomics and proteomics tools in mammalian stem cells, cancer cell lines and animal models. Our research group values inclusivity and curiosity, where we strive for a balance between creativity and scientific rigor that is essential for enabling transformative discoveries.

Projects Available in the Isbel Research Group
- Project Title: Dissecting the molecular interplay between transcription factors and chromatin
- Project Supervisor(s): Dr Luke Isbel
- Suitable for: MPhil, PhD
- Location of project: SAiGENCI, Adeliade
Project outline:
Transcription factors are proteins that bind DNA motifs in regulatory sequences, like enhancers or promoters, to activate expression of nearby genes. At the same time, the genome is embedded in chromatin, where ‘epigenetic’ marks like histone modifications are thought to provide an extra level of gene control. While we suspect there is a remarkable specificity encoded within the interplay between transcription factors and chromatin, the molecular details remain enigmatic. For example, beyond sensing the underlying DNA sequence, do transcription factors also interpret chromatin and can this affect their function?
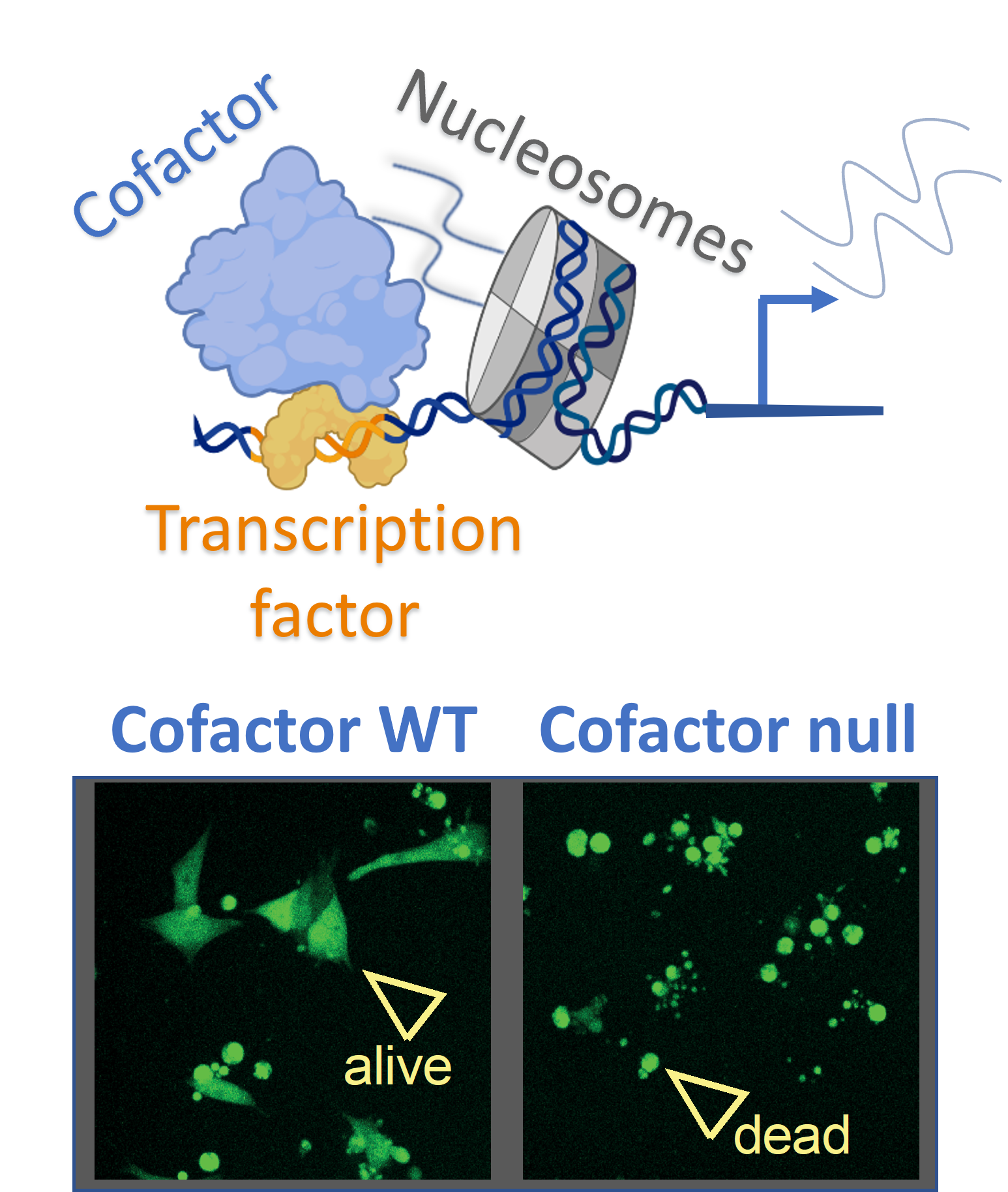
Importantly, transcription factors are often deregulated in cancer, suggesting they can be important therapeutic targets. The tumour suppressor p53 is one such transcription factor and it is frequently silenced in cancer. We recently demonstrated that specific chromatin marks can prevent p53 from activating its target genes, via a mechanism that involves a histone-binding cofactor called Trim24 (Isbel, 2023). The candidate will use this prior work as an experimental platform aimed at addressing the role of chromatin in regulating p53 function, and to link this back to the biology of stress-response in stem cells and colorectal cancer (CRC). They will establish reporters based of target genes of p53 and Trim24 which can be used to screen for additional factors involved in gene regulation, whereupon molecular tools like degron-tagging and mutant alleles will be used to investigate their function.
The ultimate goal of this project is to generate a framework for testing both transcription factors and local chromatin marks as a novel therapeutic approach in cancer.
We will fully support the candidate in learning scientific and technical skills, including a variety of genomic, epigenomic and biochemistry techniques such as CRISPR Cas9 gene-editing, flow cytometry-based genome-wide CRISPR/Cas9 screening, various genomics/epigenetic techniques (i.e. ChIPseq, ATACseq, RNAseq etc.) and sophisticated bioinformatic analysis. This will well-prepare the candidate for a future in science.
Our ideal candidate will be curious about gene regulation and demonstrate creativity in their approach. They should have some experience with basic molecular biology techniques, such as PCR, Western blotting or tissue culture etc. We would strive to recruit a candidate that is excited to shape the success of the group by bringing their own unique experiences and insights.
| Date | Institution name | Country | Title |
|---|---|---|---|
| 2012 - 2015 | La Trobe University | Australia | PhD in Molecular Genetics |
| 2011 | Griffith University | Australia | Honours, Biomedical Science |
| 2008 - 2010 | James Cook University | Australia | Bachelor of Biomedical Science |
| Year | Citation |
|---|---|
| 2025 | Liu, J., Roy, M. J., Isbel, L., & Li, F. (2025). Accurate PROTAC-targeted degradation prediction with DegradeMaster. Bioinformatics, 41(Supplement_1), i342-i351. Scopus5 WoS4 Europe PMC2 |
| 2025 | Chakraborty, D., Sandate, C. R., Isbel, L., Kempf, G., Weiss, J., Cavadini, S., . . . Thomä, N. H. (2025). Nucleosomes specify co-factor access to p53. Molecular Cell, 85(15), 2919-2936.e12. Europe PMC1 |
| 2025 | Durdu, S., Iskar, M., Isbel, L., Hoerner, L., Wirbelauer, C., Burger, L., . . . Schübeler, D. (2025). Chromatin-dependent motif syntax defines differentiation trajectories. Molecular Cell, 85(15), 2900-2918. WoS1 Europe PMC6 |
| 2023 | Isbel, L., Iskar, M., Durdu, S., Weiss, J., Grand, R. S., Hietter-Pfeiffer, E., . . . Schübeler, D. (2023). Readout of histone methylation by Trim24 locally restricts chromatin opening by p53.. Nat Struct Mol Biol, 30(7), 948-957. Scopus30 WoS33 Europe PMC33 |
| 2023 | Michael, A. K., Stoos, L., Crosby, P., Eggers, N., Nie, X. Y., Makasheva, K., . . . Thomä, N. H. (2023). Cooperation between bHLH transcription factors and histones for DNA access.. Nature, 619(7969), 385-393. Scopus67 WoS65 Europe PMC79 |
| 2023 | Yelagandula, R., Stecher, K., Novatchkova, M., Michetti, L., Michlits, G., Wang, J., . . . Bell, O. (2023). ZFP462 safeguards neural lineage specification by targeting G9A/GLP-mediated heterochromatin to silence enhancers. Nature Cell Biology, 25(1), 42-55. Scopus18 WoS17 Europe PMC20 |
| 2022 | Isbel, L., Grand, R. S., & Schübeler, D. (2022). Generating specificity in genome regulation through transcription factor sensitivity to chromatin. Nature Reviews Genetics, 23(12), 728-740. Scopus89 WoS85 Europe PMC112 |
| 2021 | Grand, R. S., Burger, L., Gräwe, C., Michael, A. K., Isbel, L., Hess, D., . . . Schübeler, D. (2021). BANP opens chromatin and activates CpG-island-regulated genes. Nature, 596(7870), 133-137. Scopus68 WoS68 Europe PMC78 |
| 2020 | Michael, A. K., Grand, R. S., Isbel, L., Cavadini, S., Kozicka, Z., Kempf, G., . . . Thomä, N. H. (2020). Mechanisms of OCT4-SOX2 motif readout on nucleosomes. Science, 368(6498), 1460-1465. Scopus168 WoS162 Europe PMC204 |
| 2019 | Hartl, D., Krebs, A. R., Grand, R. S., Baubec, T., Isbel, L., Wirbelauer, C., . . . Schübeler, D. (2019). CG dinucleotides enhance promoter activity independent of DNA methylation. Genome Research, 29(4), 554-563. Scopus41 WoS39 Europe PMC47 |
| 2019 | Russo, A. M., Lawther, A. J., Prior, B. M., Isbel, L., Somers, W. G., Lesku, J. A., . . . Hale, M. W. (2019). Social approach, anxiety, and altered tryptophan hydroxylase 2 activity in juvenile BALB/c and C57BL/6J mice. Behavioural Brain Research, 359, 918-926. Scopus14 WoS15 Europe PMC9 |
| 2018 | Isbel, L., & Schübeler, D. (2018). Non-mendelian Inheritance in Mammals Is Highly Constrained. Cell, 175(5), 1179-1181. Scopus2 WoS1 Europe PMC1 |
| 2016 | Isbel, L., Prokopuk, L., Wu, H., Daxinger, L., Oey, H., Spurling, A., . . . Whitelaw, E. (2016). Wiz binds active promoters and CTCF-binding sites and is required for normal behaviour in the mouse. Elife, 5(JULY), 39 pages. Scopus20 WoS19 Europe PMC23 |
| 2016 | Daxinger, L., Oey, H., Isbel, L., Whitelaw, N. C., Youngson, N. A., Spurling, A., . . . Whitelaw, E. (2016). Hypomethylation of ERVs in the sperm of mice haploinsufficient for the histone methyltransferase Setdb1 correlates with a paternal effect on phenotype. Scientific Reports, 6(1), 10 pages. Scopus23 WoS20 Europe PMC19 |
| 2015 | Isbel, L., Srivastava, R., Oey, H., Spurling, A., Daxinger, L., Puthalakath, H., & Whitelaw, E. (2015). Trim33 Binds and Silences a Class of Young Endogenous Retroviruses in the Mouse Testis; a Novel Component of the Arms Race between Retrotransposons and the Host Genome. Plos Genetics, 11(12), 25 pages. Scopus23 WoS22 Europe PMC24 |
| 2015 | Oey, H., Isbel, L., Hickey, P., Ebaid, B., & Whitelaw, E. (2015). Genetic and epigenetic variation among inbred mouse littermates: Identification of inter-individual differentially methylated regions. Epigenetics and Chromatin, 8(1), 12 pages. Scopus50 WoS47 Europe PMC49 |
| 2015 | Isbel, L., & Whitelaw, E. (2015). Commentary: Far-reaching hypothesis or a step too far: The inheritance of acquired characteristics. International Journal of Epidemiology, 44(4), 1109-1112. Scopus6 WoS6 Europe PMC3 |
| 2015 | Harten, S. K., Oey, H., Bourke, L. M., Bharti, V., Isbel, L., Daxinger, L., . . . Whitelaw, E. (2015). The recently identified modifier of murine metastable epialleles, Rearranged L-Myc Fusion, is involved in maintaining epigenetic marks at CpG island shores and enhancers. BMC Biology, 13(1), 15 pages. Scopus16 WoS15 Europe PMC15 |
| 2013 | Daxinger, L., Harten, S. K., Oey, H., Epp, T., Isbel, L., Huang, E., . . . Whitelaw, E. (2013). An ENU mutagenesis screen identifies novel and known genes involved in epigenetic processes in the mouse. Genome Biology, 14(9), 17 pages. Scopus69 WoS68 Europe PMC72 |
| 2012 | Isbel, L., & Whitelaw, E. (2012). Endogenous retroviruses in mammals: An emerging picture of how ervs modify expression of adjacent genes. Bioessays, 34(9), 734-738. Scopus29 WoS28 Europe PMC26 |
| Year | Citation |
|---|---|
| 2025 | Liu, J., Roy, M., Isbel, L., & Li, F. (2025). Accurate PROTAC targeted degradation prediction with DegradeMaster. DOI Europe PMC1 |
2025-2030 Viertel Foundation Senior Medical Research Fellowship.
2019 Chique-Ehrismann Originality prize, Friedrich Miescher Institute.
2019-2023 Early Career Fellowship, National Health and Medical Research Council of Australia.
2017-2019 Marie Skłodowska-Curie Individual Fellowship, European Commission.
Guest Lecturer Genetics III
| Date | Role | Research Topic | Program | Degree Type | Student Load | Student Name |
|---|---|---|---|---|---|---|
| 2026 | Principal Supervisor | Gene regulation and chromatin biology | - | Doctorate | Full Time | Mr Grant Timothy Freeman |
| 2025 | Principal Supervisor | Exploring the molecular interplay of highly related tumour suppressor proteins that bind identical DNA motifs | Master of Philosophy (Medical Science) | Master | Full Time | Miss Katarina Anastasia Amerl |
| 2025 | Principal Supervisor | Basic research in Cancer Epigenetics | Doctor of Philosophy | Doctorate | Full Time | Ms Begona Nieto Fernandez |
| 2025 | Principal Supervisor | Exploring the molecular interplay of highly related tumour suppressor proteins that bind identical DNA motifs | - | Master | Full Time | Miss Katarina Anastasia Amerl |
| 2025 | Principal Supervisor | Basic research in Cancer Epigenetics | Doctor of Philosophy | Doctorate | Full Time | Ms Begona Nieto Fernandez |



