
Prof Lan Nguyen
Program Leader, Computational Systems Oncology
SAIGENCI
College of Health
Eligible to supervise Masters and PhD - email supervisor to discuss availability.
Professor Lan Nguyen was appointed in 2025 to lead the Computational Systems Oncology Program at the South Australian immunoGENomics Cancer Institute (SAiGENCI), bringing extensive expertise in integrating advanced computational modelling with cutting-edge biological techniques to understand and combat cancer.Professor Nguyen completed his PhD in Computational Systems Biology before joining Systems Biology Ireland, where he trained under international leaders in systems biology. After advancing to junior group leader there, he was recruited to Monash Biomedicine Discovery Institute in 2015, where he established one of the few research labs worldwide combining both computational modelling and experimental capabilities under one roof.Professor Nguyen is internationally recognised for developing sophisticated, experimentally-validated computational models of important biological systems. Through exploiting systems-based approaches that synergise advanced techniques from computational and experimental sciences, his studies have established new systems-level insights into cell signalling and cell-fate determination. His interdisciplinary research has shed new light on mechanisms of anti-cancer drug resistance and identified novel therapeutic strategies. His work has appeared in high-impact journals including Nature Cell Biology, Cell Systems, eLife, Nature Communications, and Cancer Research. He has secured more than $5.5 million in competitive CIA funding and serves as a Chief Investigator of the ARC Centre of Excellence in Mathematical Modelling of Cellular Systems (MACSYS).His innovative approaches have earned multiple honours, including a Victorian Cancer Agency Mid-Career Fellowship, CIA of a prestigious Cancer Council Victoria Venture Grant, and editorship of an authoritative book on computational modelling published by Nature Springer. At SAiGENCI, Professor Nguyen aims to advance cancer research through quantitative and predictive approaches while developing computational models that will serve as powerful decision-support tools for personalised cancer treatment.
WELCOME TO THE NGUYEN LAB
The goal of our lab is to gain network-level insights into the innerworkings of human cells under normal and cancer contexts, through development and exploitation of innovative systems-based approaches that integrate mathematical modelling and computational simulation, AI, bioinformatics with in-house wet lab experiment.
Research vision
Decades of experimental work using traditional biochemical approaches have been highly successful in characterising the function of single genes and proteins, and mapping them into functional pathways. However, it has become abundantly clear that cell signalling pathways do not act in isolation but interlink and cooperate to direct how cells respond to intra/extra-cellular cues, enabled by a multitude of complex crosstalk and feedback mechanisms. This realization together with the advent of -omics technologies that provide the ability to routinely measure thousands of genes and proteins, have necessitated a shift from studying single molecules to the study of interconnected networks. To fully understand cell signalling and behaviour, we thus need to move beyond studying individual pathways in isolation and undertake systematic and quantitative analysis of integrated networks that link functionally related pathways. This is, however, challenging to achieve via the historical and pathway-centric approach; instead, computational network modelling in integration with experimental analysis represents an innovative and powerful strategy to characterise biochemical networks, understand their dynamic behaviours and fully exploit their therapeutic potential.
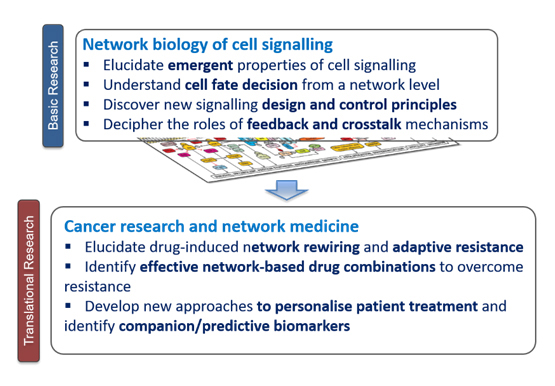
Figure 1. Primary research themes currently undertaken in the Nguyen laboratory.
Driven by this ‘systems biology’ paradigm, the primary interest of the Nguyen Lab is to develop and exploit innovative systems-based approaches that integrate predictive computational modelling, bioinformatic techniques and wet-lab experiments to address fundamental questions in cell biology and cancer fields (Figure 1). A central feature of our work is the highly interdisciplinary and iterative nature of the approaches (Figure 2), which involve data generation, model building and training, model analysis and prediction, and experimental validation. The Nguyen lab thus nourishes a strongly cross-disciplinary research environment consisting of mixed expertise and resources.
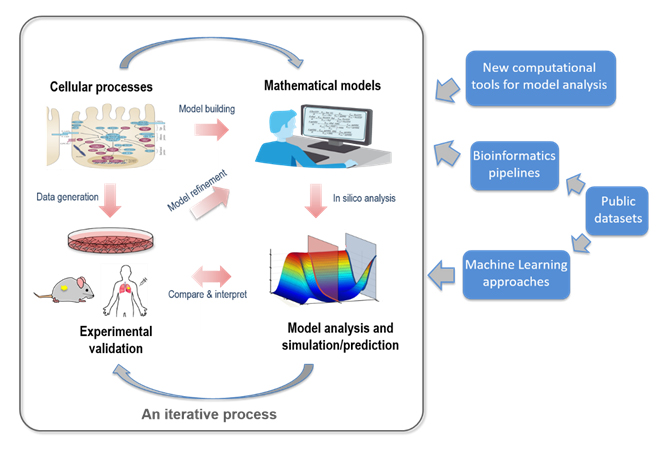
Figure 2. An integrative modelling-experimental approach central to our studies.
Selective projects
Below are description of some selective projects ongoing in the lab - please contact Prof Nguyen for a full list.
- Tackling adaptive resistance to anti-cancer drugs using systems modelling approach
- Friend or foe: Deciphering the conflicting functions of YAP in cancer
- Mathematical modelling of the PI3K-Akt-mTOR network
- Developing novel analytical and visualisation techniques for model analysis
- Network-level characterisation of TCR signalling in T cells
1. Tackling adaptive resistance to anti-cancer drugs using systems modelling approach
Targeted therapy has revolutionised cancer treatment, yet development of resistance to these therapies remains a primary reason for treatment failure and patient relapse. To date, studies have been largely focused on elucidating the genetic mechanisms of drug resistance. However, it has become clear that tumour cells also rely on non-genetic and highly adaptive mechanisms involving rapid and dynamic rewiring of signalling pathways in order to bypass the drug blockade. A key reason underlying this phenomenon is the complexity and highly interconnected nature of cell signalling pathways, featuring abundant crosstalk and feedback regulation that have evolved to provide homeostasis and robustness to normal cells, but are hijacked by transformed cells to overcome anti-cancer drug agents. Overcoming adaptive resistance requires an ability to integrate multiple cancer-relevant signalling pathways in unified frameworks, and describe their dynamics quantitatively at a system level.
Over the past five years, my laboratory has successfully exploited this integrative approach to gain new important systems-level insights into oncogenic signalling networks, placing our research at the forefront of the field. Exploiting this competitive position, this research theme aims to undertake a systems-level characterisation of drug-induced network remodelling; and based on this understanding, to devise new effective combination therapies for cancer. This will be achieved via an integrated approach that synergises predictive modelling and cutting-edge lab-based experiment (Figure 3).
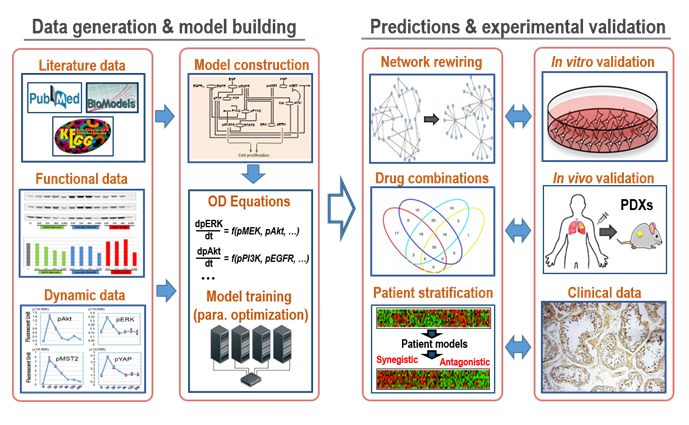
Figure 3. A workflow of our systems-based approach to drug combination discovery.
1a. Overcoming network rewiring-mediated resistance in aggressive breast cancer
Triple-negative breast cancer (TNBC) is highly aggressive but patients have no targeted treatment options beside toxic chemotherapies. Like ants who are very good at seeking alternative ways to travel when their path is blocked, TNBC cells manage to find escape paths to bypass drug treatment. Combining computer simulations and lab work, this project aims to predict these escape paths and identify ways to block them by combining multiple drugs, which will lead to new treatments for this disease. Specifically, we are building predictive computational models of specific signalling networks in TNBC and will apply our recently developed Synergistic Drug Combination Discovery (SynDISCO) framework (PLOS Comp Biol, 2018) where the co-inhibition effects of all possible pair-wise combinations of network nodes are systematically evaluated, and compared, based on well-established quantitative drug synergy metrics. This provides a rational approach for ranking and prioritising drug combinations, thereby greatly enhancing the efficiency of therapeutic strategy development. Prioritized drug combinations will then be tested in appropriate in vitro and in vivo models. This work is being undertaken in collaboration with Professor Roger Daly (Monash University, Melbourne), Associate Professor Alex Swarbrick (Garvan Institute of Medical Research, Sydney) and clinician oncologist Professor Sherene Loi (Peter McCallum Cancer Centre).
1b. Unravelling mechanisms of resistance to PI3K inhibitors through integrative modelling of signalling network
The PI3K/Akt pathway plays a vital role in orchestrating multiple cellular responses such as metabolism, proliferation and survival. This pathway is highly activated in primary human cancers including breast cancer. PIK3CA, the gene coding for catalytic subunit p110α of PI3K, is the most commonly mutated oncogene in breast cancer. This implies that selective inhibition of PI3Kα may have robust antitumor efficacy in PIK3CA-mutant cancers, however, clinical responses to PI3Kα specific inhibitors as single-agent therapy have been disappointing due to intrinsic and acquired resistance. In collaboration with Dr Antonella Papa (Monash University), we are constructing novel multi-pathway computational models of PI3K signalling in combination with experimental validation to investigate network-level mechanisms of resistance to PI3Kα inhibition, based on which to identify drug combinations that can overcome this resistance.
2. Friend or foe: Deciphering the conflicting functions of YAP in cancer
The Hippo/YAP signalling pathway is dysregulated in ~80% of breast cancer cases, making it a potential therapeutic target. However, ongoing efforts aimed at targeting this pathway have yielded limited success. This is due to the fact that YAP, the pathway’s downstream effector and most feasible drug target, is found to display either pro-growth or pro-death function depending on specific contexts. Currently, it is not understood when and how YAP exhibits which function, and this lack of understanding hampers translation of YAP-directed therapy into clinics. Our data and others have indicated that the specific function of YAP is driven by its ability to form functionally distinct complexes with various transcriptional factors, and such formations are tightly controlled by phosphorylation events. This project utilises integrative systems biology approaches combining predictive computational modelling with experiments to decipher the dual functions of YAP in breast cancer, and exploit that understanding to develop effective ways to target YAP.
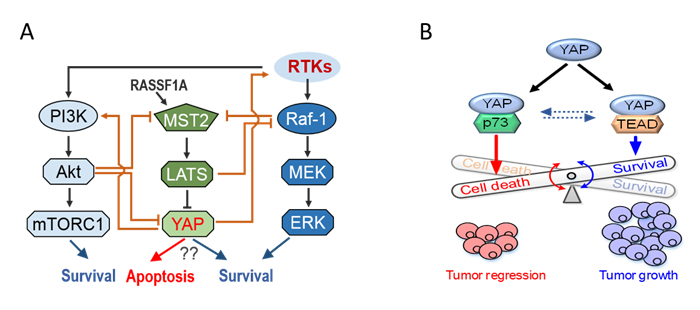
Figure 4. (A) A simplified diagram depicting the YAP regulatory network. (B) A schematic depicting the dual role of YAP in cancer regulation. Cancer cells tipped towards survival when YAP binds TEAD.
3. Mathematical modelling of the PI3K-Akt-mTOR network
3a. Identifying improved treatments targeting the PI3K pathway
The PI3K-Akt-mTOR signalling network plays a pivotal role in the regulation of cell growth and proliferation, and is highly complex with multiple feedback loops, pathway crosstalk, upstream regulators and downstream functions. Its frequent aberrations in cancers makes this network an important therapeutic target, and indeed many targeted drugs have been developed directed at its components. However, the clinical success in inhibiting PI3K, Akt, mTORC1 and/or mTORC2 has been disappointing due to the emergence of drug resistance. This project will employ an integrative systems approach to gain systems-level understanding of the network dynamics in cancer cells before and after drug treatment. This knowledge together with model simulations will help design new therapeutic strategies that exploit network vulnerabilities and are capable of overcoming resistance. Predictions will then be tested experimentally in the wet lab.
3b. Elucidating the functional role of DEPTOR in cancer and exploiting it for therapeutic purposes
DEPTOR was discovered as a unique endogenous inhibitor of both mTOR Complex 1 and 2. Intriguingly, DEPTOR expression is highly variable across various cancer types, and the functional role of DEPTOR in many cancer remains conflicting and poorly understood. We have recently constructed the first mathematical model of the mTOR/DEPTOR network and demonstrated a critical role of DEPTOR in regulating the network’s complex dynamic behaviour (Figure 5). This project continues to characterise the functional role of DEPTOR in cancer, with an aim to design effective therapeutic strategy targeting this protein.
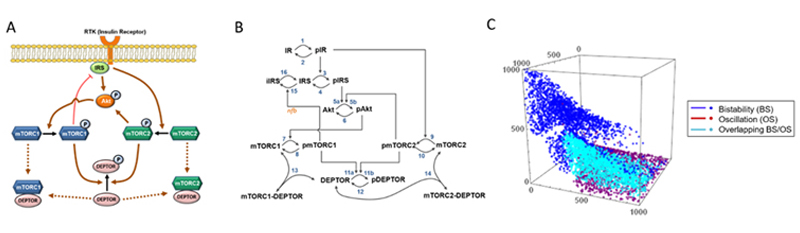
Figure 5. (A) Simplified diagram depicting the interactions and feedback loops within the DEPTOR-mTOR network. (B) Reaction scheme of the DEPTOR-mTOR mathematical model. (C) 3D simulation showing the effect of chaning model state variables on network dynamical behaviours, demonstrating a critical role of DEPTOR in modulating network dynamics.
4. Developing novel analytical and visualisation techniques for model analysis
Biochemical networks are dynamic and multi-dimensional systems, consisting of tens or hundreds of molecular components. Elucidating the network dynamics in health and disease is crucial to better understand the disease mechanisms and derive effective therapeutic strategies. However, current approaches to analyse and visualise systems dynamics can often provide only low-dimensional projections of the network dynamics, which often does not present the multi-dimensional picture of the system behaviour. To address this issue, we have developed an innovative visualisation framework for high-dimensional network behaviour named “Dynamics Visualisation based on Parallel Coordinates” (DYVIPAC) which exploits the advantages provided by parallel coordinates graphs. DYVIPAC has proven to be very powerful in elucidating the governing conditions underlying specific network behaviours that are hidden in the high-dimensional space. This project aims to extend DYVIPAC by providing a GUI to this framework, in order to provide better and user-friendly ways to analyse and visualise complex network behaviours.
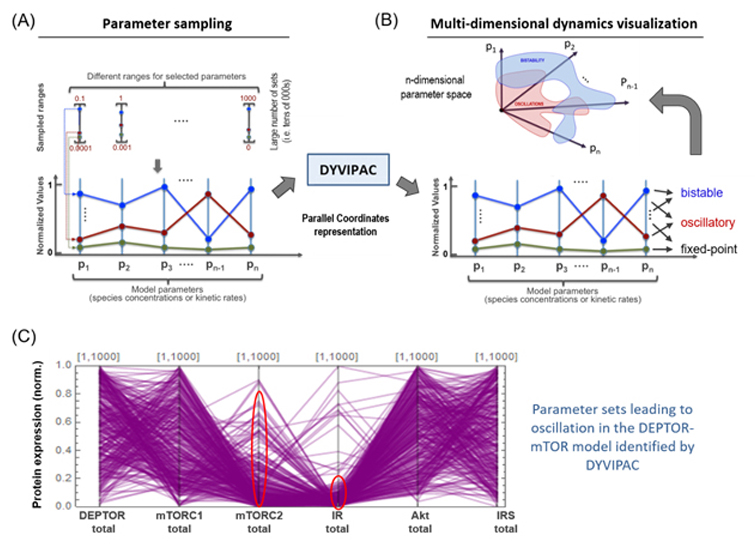
Figure 6. (A-B) Schematic illustrating DYVIPAC-based multi-dimensional analysis of dynamic behaviours and visualisation using Parallel Coordinates (PC) plots (see Nguyen et al. (2015) for details). (C) PC plot showing the oscillations-inducing sets returned from a 6D analysis applied to the DEPTOR-mTOR model above.
5. Network-level characterisation of TCR signalling in T cells
A properly functioning immune system is critical for an organism’s health. Immune tolerance is a critical feature of the immune system that allows immune cells to mount effective responses against exogenous pathogens such as viruses and bacteria, while preventing attack to self-tissues. Activation-induced cell death (AICD) in T lymphocytes, in which repeated stimulations of the T-cell receptor (TCR) lead to activation and then apoptosis of T cells, is a major mechanism for T cell homeostasis and helps to maintain peripheral immune tolerance. A defect in AICD may lead to development of autoimmune diseases. Despite its importance, the regulatory mechanisms that underlie AICD remain poorly understood particularly at a systems level. In this project, we develop a mechanistic, multi-pathway, and dynamic model of the TCR signalling network and perform computational analyses to characterize the salient systems-level properties of AICD.
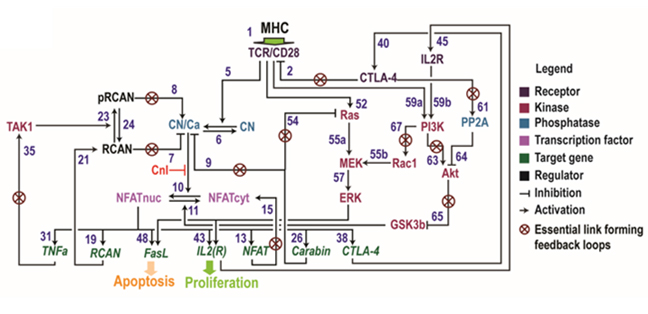
Figure 7. Schematic of a mathematical network model integrating multiple signalling pathways downstream of the T cell receptor (Shin et al. 2019).
Techniques/expertise
Kinetic, mechanistic and dynamic pathway modelling and simulation (eg MATLAB, Mathematica)
Ordinary Differential Equation (ODE) modelling
Agent-based modelling
Sophisticated model calibration and parameter estimation
Model-based perturbation and sensitivity analysis
Systems dynamical analysis and visualisation
Immunoblotting, RPPA
CRISPR, siRNA, cell imaging
Cellular assays (eg cell viability, apoptosis, migration, colono formation)
RNA-seq, Phosphoproteomics
| Date | Position | Institution name |
|---|---|---|
| 2024 - ongoing | Program Lead, Computational Systems Oncology | University of Adelaide |
| 2024 - ongoing | Professor | University of Adelaide |
| 2024 - ongoing | Head, Integrative Network Modelling Lab | University of Adelaide |
| 2021 - 2024 | Associate Professor | Monash University |
| 2019 - 2023 | Victoria Cancer Agency Mid-Career Research Fellow | Monash University |
| 2015 - ongoing | Head, Integrative Network Modelling Lab | Monash University |
| 2014 - 2015 | Junior Group Leader | University College Dublin |
| 2010 - 2014 | Postdoctoral Research Fellow | University College Dublin |
| Date | Institution name | Country | Title |
|---|---|---|---|
| 2007 - 2010 | Lincoln University | New Zealand | PhD |
| 2001 - 2005 | Lincoln University | New Zealand | B Appl Maths & Computing (Hons) |
| Year | Citation |
|---|---|
| 2025 | Yang, X., Ma, X., Zhao, T., Croucher, D. R., Nguyen, E. V., Clark, K. C., . . . Daly, R. J. (2025). Activation of CAMK2 by pseudokinase PEAK1 represents a targetable pathway in triple negative breast cancer. Nature Communications, 16(1), 1871-1-1871-19. Scopus2 WoS3 Europe PMC3 |
| 2025 | Lan, Y., Shin, S. Y., & Nguyen, L. K. (2025). From shallow to deep: The evolution of machine learning and mechanistic model integration in cancer research. Current Opinion in Systems Biology, 40, 11 pages. Scopus5 WoS5 |
| 2024 | Shin, S. Y., Chew, N. J., Ghomlaghi, M., Chüeh, A. C., Jeong, Y., Nguyen, L. K., & Daly, R. J. (2024). Integrative Modeling of Signaling Network Dynamics Identifies Cell Type–Selective Therapeutic Strategies for FGFR4-Driven Cancers. Cancer Research, 84(19), 3296-3309. Scopus4 WoS4 Europe PMC1 |
| 2024 | Yip, H. Y. K., Shin, S. Y., Chee, A., Ang, C. S., Rossello, F. J., Wong, L. H., . . . Papa, A. (2024). Integrative modeling uncovers p21-driven drug resistance and prioritizes therapies for PIK3CA-mutant breast cancer. npj Precision Oncology, 8(1), 20-1-20-21. Scopus14 WoS12 Europe PMC8 |
| 2024 | Ghomlaghi, M., Theocharous, M., Hoang, N., Shin, S. Y., von Kriegsheim, A., O’ Neill, E., . . . Nguyen, L. K. (2024). Integrative modeling and analysis of signaling crosstalk reveal molecular switches coordinating Yes-associated protein transcriptional activities. iScience, 27(3), 109031-1-109031-19. Scopus7 WoS6 Europe PMC6 |
| 2024 | Didan, Y., Ghomlaghi, M., Nguyen, L. K., & Ng, D. C. H. (2024). Stress pathway outputs are encoded by pH-dependent clustering of kinase components.. Nature Communications, 15(1), 6614-1-6614-16. Scopus3 WoS3 Europe PMC4 |
| 2023 | Shin, S. Y., Centenera, M. M., Hodgson, J. T., Nguyen, E. V., Butler, L. M., Daly, R. J., & Nguyen, L. K. (2023). A Boolean-based machine learning framework identifies predictive biomarkers of HSP90-targeted therapy response in prostate cancer. Frontiers in Molecular Biosciences, 10, 1094321-1-1094321-16. Scopus7 WoS6 Europe PMC5 |
| 2023 | Yang, X., Cruz, M. I., Nguyen, E. V., Huang, C., Schittenhelm, R. B., Luu, J., . . . Daly, R. J. (2023). The pseudokinase NRBP1 activates Rac1/Cdc42 via P-Rex1 to drive oncogenic signalling in triple-negative breast cancer. Oncogene, 42(11), 833-847. Scopus10 WoS10 Europe PMC9 |
| 2021 | Kearney, A. L., Norris, D. M., Ghomlaghi, M., Wong, M. K. L., Humphrey, S. J., Carroll, L., . . . Burchfield, J. G. (2021). Akt phosphorylates insulin receptor substrate to limit pi3k-mediated pip3 synthesis. eLife, 10, e66942. Scopus66 Europe PMC49 |
| 2021 | Norris, D., Yang, P., Shin, S. Y., Kearney, A. L., Kim, H. J., Geddes, T., . . . Burchfield, J. G. (2021). Signaling Heterogeneity is Defined by Pathway Architecture and Intercellular Variability in Protein Expression. iScience, 24(2), 102118. Scopus14 |
| 2021 | Ghomlaghi, M., Yang, G., Shin, S., James, D. E., & Nguyen, L. K. (2021). Dynamic modelling of the PI3K/MTOR signalling network uncovers biphasic dependence of mTORC1 activity on the mTORC2 subunit SIN1. PLoS Computational Biology, 17(9), 26 pages. Scopus14 WoS12 Europe PMC14 |
| 2021 | Ghomlaghi, M., Hart, A., Hoang, N., Shin, S., & Nguyen, L. K. (2021). Feedback, crosstalk and competition: Ingredients for emergent non‐linear behaviour in the pi3k/mtor signalling network. International Journal of Molecular Sciences, 22(13), 20 pages. Scopus22 WoS20 Europe PMC26 |
| 2020 | Yip, H. Y. K., Chee, A., Ang, C. S., Shin, S. Y., Ooms, L. M., Mohammadi, Z., . . . Papa, A. (2020). Control of Glucocorticoid Receptor Levels by PTEN Establishes a Failsafe Mechanism for Tumor Suppression. Molecular Cell, 80(2), 279-295.e8. Scopus19 WoS18 Europe PMC19 |
| 2019 | Shin, S. Y., Kim, M. W., Cho, K. H., & Nguyen, L. K. (2019). Coupled feedback regulation of nuclear factor of activated T-cells (NFAT) modulates activation-induced cell death of T cells. Scientific Reports, 9(1), 15 pages. Scopus12 WoS13 Europe PMC13 |
| 2019 | Su, Z., Burchfield, J. G., Yang, P., Humphrey, S. J., Yang, G., Francis, D., . . . James, D. E. (2019). Global redox proteome and phosphoproteome analysis reveals redox switch in Akt. Nature Communications, 10(1), 18 pages. Scopus118 WoS112 Europe PMC107 |
| 2019 | Padman, B. S., Nguyen, T. N., Uoselis, L., Skulsuppaisarn, M., Nguyen, L. K., & Lazarou, M. (2019). LC3/GABARAPs drive ubiquitin-independent recruitment of Optineurin and NDP52 to amplify mitophagy. Nature Communications, 10(1), 13 pages. Scopus195 WoS188 Europe PMC190 |
| 2018 | Shin, S. Y., Müller, A. K., Verma, N., Lev, S., & Nguyen, L. K. (2018). Systems modelling of the EGFR-PYK2-c-Met interaction network predicts and prioritizes synergistic drug combinations for triple-negative breast cancer. Plos Computational Biology, 14(6), 30 pages. Scopus28 WoS21 Europe PMC19 |
| 2017 | Verma, N., Müller, A. K., Kothari, C., Panayotopoulou, E., Kedan, A., Selitrennik, M., . . . Lev, S. (2017). Targeting of PYK2 synergizes with EGFR antagonists in basal-like TNBC and circumvents HER3-associated resistance via the NEDD4-NDRG1 axis. Cancer Research, 77(1), 86-99. Scopus65 WoS64 Europe PMC64 |
| 2016 | Shin, S. Y., & Nguyen, L. K. (2016). Unveiling hidden dynamics of hippo signalling: A systems analysis. Genes, 7(8), 15 pages. Scopus16 WoS15 Europe PMC15 |
| 2016 | Zhang, Y., Shen, K., Bai, Y., Lv, X., Huang, R., Zhang, W., . . . Yao, H. (2016). <i>Mir143</i>-BBC3 cascade reduces microglial survival via interplay between apoptosis and autophagy: Implications for methamphetamine-mediated neurotoxicity. AUTOPHAGY, 12(9), 1538-1559. WoS49 Europe PMC45 |
| 2016 | Nguyen, L. K. (2016). Dynamics of ubiquitin-mediated signalling: insights from mathematical modelling and experimental studies. BRIEFINGS IN BIOINFORMATICS, 17(3), 479-493. WoS9 Europe PMC9 |
| 2016 | Fabian, Z., Taylor, C. T., & Nguyen, L. K. (2016). Understanding complexity in the HIF signaling pathway using systems biology and mathematical modeling. JOURNAL OF MOLECULAR MEDICINE-JMM, 94(4), 377-390. WoS21 Europe PMC19 |
| 2016 | Rodriguez, J., Pilkington, R., Munoz, A. G., Nguyen, L. K., Rauch, N., Kennedy, S., . . . von Kriegsheim, A. (2016). Substrate-Trapped Interactors of PHD3 and FIH Cluster in Distinct Signaling Pathways. CELL REPORTS, 14(11), 2745-2760. WoS75 Europe PMC71 |
| 2016 | Nguyen, L. K., & Kholodenko, B. N. (2016). Feedback regulation in cell signalling: Lessons for cancer therapeutics. SEMINARS IN CELL & DEVELOPMENTAL BIOLOGY, 50, 85-94. WoS49 Europe PMC51 |
| 2016 | Byrne, K. M., Monsefi, N., Dawson, J. C., Degasperi, A., Bukowski-Wills, J. -C., Volinsky, N., . . . Kholodenko, B. N. (2016). Bistability in the Rac1, PAK, and RhoA Signaling Network Drives Actin Cytoskeleton Dynamics and Cell Motility Switches. CELL SYSTEMS, 2(1), 38-48. WoS134 Europe PMC134 |
| 2016 | Jambrina, P. G., Rauch, N., Pilkington, R., Rybakova, K., Nguyen, L. K., Kholodenko, B. N., . . . Rosta, E. (2016). Phosphorylation of RAF Kinase Dimers Drives Conformational Changes that Facilitate Transactivation. ANGEWANDTE CHEMIE-INTERNATIONAL EDITION, 55(3), 983-986. WoS39 Europe PMC41 |
| 2016 | Shen, K., Zhang, Y., Lv, X., Chen, X., Zhou, R., Nguyen, L. K., . . . Yao, H. (2016). Molecular Mechanisms Involving Sigma-1 Receptor in Cell Apoptosis of BV-2 Microglial Cells Induced by Methamphetamine. CNS & NEUROLOGICAL DISORDERS-DRUG TARGETS, 15(7), 857-865. WoS16 |
| 2016 | Weitsman, G., Barber, P. R., Nguyen, L. K., Lawler, K., Patel, G., Woodman, N., . . . Ng, T. (2016). HER2-HER3 dimer quantification by FLIM-FRET predicts breast cancer metastatic relapse independently of HER2 IHC status. ONCOTARGET, 7(32), 51012-51026. WoS29 |
| 2016 | Sanchez-Sanz, G., Matallanas, D., Nguyen, L. K., Kholodenko, B. N., Rosta, E., Kolch, W., & Buchete, N. -V. (2016). MST2-RASSF protein-protein interactions through SARAH domains. BRIEFINGS IN BIOINFORMATICS, 17(4), 593-602. WoS10 Europe PMC13 |
| 2015 | Varusai, T. M., Kolch, W., Kholodenko, B. N., & Nguyen, L. K. (2015). Protein-protein interactions generate hidden feedback and feed-forward loops to trigger bistable switches, oscillations and biphasic dose-responses. MOLECULAR BIOSYSTEMS, 11(10), 2750-2762. WoS26 Europe PMC23 |
| 2015 | Nguyen, L. K., Cavadas, M. A. S., Kholodenko, B. N., Frank, T. D., & Cheong, A. (2015). Species differential regulation of COX2 can be described by an NFκB-dependent logic AND gate. CELLULAR AND MOLECULAR LIFE SCIENCES, 72(12), 2431-2443. WoS18 Europe PMC14 |
| 2015 | Nguyen, L. K., Cavadas, M. A. S., Scholz, C. C., Fitzpatrick, S. F., Bruning, U., Cummins, E. P., . . . Cheong, A. (2015). A dynamic model of the hypoxia-inducible factor 1a (HIF-1a) network (vol 126, pg 1454, 2013). JOURNAL OF CELL SCIENCE, 128(2), 422. WoS10 Europe PMC11 |
| 2015 | Nguyen, L. K., Matallanas, D. G., Romano, D., Kholodenko, B. N., & Kolch, W. (2015). Competing to coordinate cell fate decisions: the MST2-Raf-1 signaling device. CELL CYCLE, 14(2), 189-199. WoS24 Europe PMC24 |
| 2015 | Nguyen, L. K., Degasperi, A., Cotter, P., & Kholodenko, B. N. (2015). DYVIPAC: an integrated analysis and visualisation framework to probe multi-dimensional biological networks. SCIENTIFIC REPORTS, 5(1), 17 pages. WoS17 Europe PMC21 |
| 2015 | Prencipe, M., O'Neill, A., O'Hurley, G., Nguyen, L. K., Fabre, A., Bjartell, A., . . . Watson, R. W. (2015). Relationship Between Serum Response Factor and Androgen Receptor in Prostate Cancer. PROSTATE, 75(15), 1704-1717. WoS7 Europe PMC6 |
| 2014 | Nguyen, L. K., Zhao, Q., Varusai, T. M., & Kholodenko, B. N. (2014). Ubiquitin chain specific auto-ubiquitination triggers sustained oscillation, bistable switches and excitable firing. IET SYSTEMS BIOLOGY, 8(6), 282-292. WoS6 Europe PMC8 |
| 2014 | den Breems, N. Y., Nguyen, L. K., & Kulasiri, D. (2014). Integrated signaling pathway and gene expression regulatory model to dissect dynamics of <i>Escherichia coli</i> challenged mammary epithelial cells. BIOSYSTEMS, 126, 27-40. WoS7 Europe PMC4 |
| 2014 | Dobrzynski, M., Nguyen, L. K., Birtwistle, M. R., von Kriegsheim, A., Fernandez, A. B., Cheong, A., . . . Kholodenko, B. N. (2014). Nonlinear signalling networks and cell-to-cell variability transform external signals into broadly distributed or bimodal responses. JOURNAL OF THE ROYAL SOCIETY INTERFACE, 11(98), 10 pages. WoS21 Europe PMC20 |
| 2014 | Kiuchi, T., Ortiz-Zapater, E., Monypenny, J., Matthews, D. R., Nguyen, L. K., Barbeau, J., . . . Ng, T. (2014). The ErbB4 CYT2 variant protects EGFR from ligand-induced degradation to enhance cancer cell motility. SCIENCE SIGNALING, 7(339), 14 pages. WoS32 Europe PMC30 |
| 2014 | Romano, D., Nguyen, L. K., Matallanas, D., Halasz, M., Doherty, C., Kholodenko, B. N., & Kolch, W. (2014). Protein interaction switches coordinate Raf-1 and MST2/Hippo signalling. NATURE CELL BIOLOGY, 16(7), 673-+. WoS122 Europe PMC130 |
| 2014 | Nguyen, L. K., Dobrzynski, M., Fey, D., & Kholodenko, B. N. (2014). Polyubiquitin chain assembly and organization determine the dynamics of protein activation and degradation. FRONTIERS IN PHYSIOLOGY, 5, 13 pages. WoS23 Europe PMC22 |
| 2013 | Nguyen, L. K., Cavadas, M. A. S., Scholz, C. C., Fitzpatrick, S. F., Bruning, U., Cummins, E. P., . . . Cheong, A. (2013). A dynamic model of the hypoxia-inducible factor 1α (HIF-1α) network. JOURNAL OF CELL SCIENCE, 126(6), 1454-1463. WoS101 Europe PMC93 |
| 2013 | Nguyen, L. K., Matallanas, D., Croucher, D. R., von Kriegsheim, A., & Kholodenko, B. N. (2013). Signalling by protein phosphatases and drug development: a systems-centred view. FEBS JOURNAL, 280(2), 751-765. WoS42 Europe PMC37 |
| 2013 | Cavadas, M. A. S., Nguyen, L. K., & Cheong, A. (2013). Hypoxia-inducible factor (HIF) network: insights from mathematical models. CELL COMMUNICATION AND SIGNALING, 11(1), 16 pages. WoS64 Europe PMC54 |
| 2013 | Nguyen, L. K., Kolch, W., & Kholodenko, B. N. (2013). When ubiquitination meets phosphorylation: a systems biology perspective of EGFR/MAPK signalling. CELL COMMUNICATION AND SIGNALING, 11(1), 15 pages. WoS157 Europe PMC155 |
| 2012 | Nguyen, L. K. (2012). Regulation of oscillation dynamics in biochemical systems with dual negative feedback loops. JOURNAL OF THE ROYAL SOCIETY INTERFACE, 9(73), 1998-2010. WoS26 Europe PMC27 |
| 2011 | Nguyen, L. K., Munoz-Garcia, J., Maccario, H., Ciechanover, A., Kolch, W., & Kholodenko, B. N. (2011). Switches, Excitable Responses and Oscillations in the Ring1B/Bmi1 Ubiquitination System. PLOS COMPUTATIONAL BIOLOGY, 7(12), 11 pages. WoS30 Europe PMC31 |
| 2011 | Nguyen, L. K., & Kulasiri, D. (2011). Distinct noise-controlling roles of multiple negative feedback mechanisms in a prokaryotic operon system. IET SYSTEMS BIOLOGY, 5(2), 145-U123. WoS4 Europe PMC3 |
| 2009 | Nguyen, L. K., & Kulasiri, D. (2009). On the functional diversity of dynamical behaviour in genetic and metabolic feedback systems. BMC SYSTEMS BIOLOGY, 3(1), 30 pages. WoS16 Europe PMC17 |
| 2008 | Nguyen, L. K., & Kulasiri, D. (2008). On multiple regulatory mechanisms in the tryptophan operon system in Escherichia coli: in silico study of perturbation dynamics.. In silico biology, 8(5-6), 485-510. Europe PMC2 |
| 2008 | Kulasiri, D., Nguyen, L. K., Samarasinghe, S., & Xie, Z. (2008). A Review of Systems Biology Perspective on Genetic Regulatory Networks with Examples. CURRENT BIOINFORMATICS, 3(3), 197-225. WoS23 |
| Year | Citation |
|---|---|
| 2014 | Weitsman, G., Barber, P. R., Lawler, K., Gillett, C., Woodman, N., Kholodenko, B., . . . Ng, T. (2014). Her2-3 heterodimer is a new and better than HER2 IHC score for clinical outcome prognosis. In EUROPEAN JOURNAL OF CANCER Vol. 50 (pp. 77). SPAIN, European Org Res & Treatment Canc, Barcelona: ELSEVIER SCI LTD. DOI |
| 2007 | Nguyen, L. K., & Kulasiri, D. (2007). Effect of Molecular Noise on the Dynamics of Tryptophan Operon System in <i>Escherichia coli</i>. In L. Oxley, & D. Kulasiri (Eds.), MODSIM 2007: INTERNATIONAL CONGRESS ON MODELLING AND SIMULATION (pp. 2006-2012). NEW ZEALAND, Christchurch: MODELLING & SIMULATION SOC AUSTRALIA & NEW ZEALAND INC. |
| Year | Citation |
|---|---|
| 2024 | Yang, X., Ma, X., Zhao, T., Croucher, D. R., Nguyen, E. V., Clark, K. C., . . . Daly, R. J. (2024). Feed-forward stimulation of CAMK2 by the oncogenic pseudokinase PEAK1 generates a therapeutically "actionable" signalling axis in triple negative breast cancer.. DOI |
| 2024 | Xu, Z., Wu, J., Shin, S. -Y., & Nguyen, L. (2024). GeneSurv: Multi-Gene Cancer Survival Analysis Tool. DOI Europe PMC1 |
| 2024 | Zhang, T., Shin, S. -Y., McCrimmon, C., Theocharous, M., Schittenhelm, R., Jarde, T., . . . Nguyen, L. (2024). Ribosomal Biogenesis Hyperactivation and ErbB signalling Mediated Network Rewiring Causes Adaptive Resistance to FGFR2 Inhibition. DOI |
| 2023 | Shin, S. -Y., & Nguyen, L. (2023). SynDISCO: A Mechanistic Modeling-Based Framework for Predictive Prioritization of Synergistic Drug Combinations Directed at Cell Signaling Networks. DOI |
| 2023 | Hart, A., Shin, S. -Y., & Nguyen, L. (2023). Systematic Analysis of Network-driven Adaptive Resistance to CDK4/6 and Estrogen Receptor Inhibition using Meta-Dynamic Network Modelling. DOI |
| 2023 | Hart, A., Shin, S. -Y., & Nguyen, L. K. (2023). Systematic Analysis of Network-driven Adaptive Resistance to CDK4/6 and Estrogen Receptor Inhibition using Meta-Dynamic Network Modelling. DOI |
| 2023 | Hart, A., Shin, S. -Y., & Nguyen, L. K. (2023). Systematic Analysis of Network-driven Adaptive Resistance to CDK4/6 and Estrogen Receptor Inhibition using Meta-Dynamic Network Modelling. DOI |
| 2022 | Ghomlaghi, M., Theocharous, M., Shin, S. -Y., O’ Neill, E., & Nguyen, L. (2022). Systems modelling of TGF-β/Hippo signalling crosstalk uncovers molecular switches that coordinate YAP transcriptional complexes (submitted to<i>iScience</i>). DOI |
| 2020 | Ghomlaghi, M., Shin, S., Yang, G., James, D., & Nguyen, L. (2020). Dynamic modelling of the PI3K/mTOR signalling network uncovers biphasic dependence of mTORC1 activation on the mTORC2 subunit Sin1. DOI |
2025 Pioneering Grant, Tour De Cure, $50,000 (CIA)
2024-2026 Investigator Initiated Project Grant, National Breast Cancer Foundation, $800,000 (CIA)
2023-2029 MACSYS: ARC Centre of Excellence for the Mathematical Analysis of Cellular Systems, Australian Research Council, $35,000,000 (Chief Investigator, Monash Node)
2021-2023 ARC Discovery Project DP21, Australian Research Council, $421,362 (CIA)
2021-2022 ARC Linkage Infrastructure, Equipment and Facilities (LIEF) grant, $900,000 (co-CI)
2020-2022 Investigator Initiated Research Scheme, National Breast Cancer Foundation, $650,000 (CIA)
2020-2021 Venture Grant, Cancer Council Victoria, $480,000 (CIA)
2019-2023 Mid-Career Research Fellowship, Victorian Cancer Agency, $600,000 (CIA)
2019-2021 ARC Discovery Project DP19, Australian Research Council, $380,000 (CIB)
2022-2023 Project grant, Tour De Cure, $50,000 (co-CI, my postdoc Milad Ghomlaghi is CIA)
2021 Project grant, CASS Foundation (co-CI, my postdoc Sungyoung Shin is CIA)
| Date | Role | Research Topic | Program | Degree Type | Student Load | Student Name |
|---|---|---|---|---|---|---|
| 2025 | Principal Supervisor | Cancer Systems Biology | Doctor of Philosophy | Doctorate | Full Time | Mr Taha Ozair Osman |
| 2025 | Principal Supervisor | Systems Biology Integration in Cancer Therapeutics: Advancing Dynamic Prediction for Precision Medicine | Doctor of Philosophy | Doctorate | Full Time | Mr Zikang Xu |
| 2025 | Principal Supervisor | Systems Biology Integration in Cancer Therapeutics: Advancing Dynamic Prediction for Precision Medicine | Doctor of Philosophy | Doctorate | Full Time | Mr Zikang Xu |
| 2025 | Principal Supervisor | Cancer Systems Biology | Doctor of Philosophy | Doctorate | Full Time | Mr Taha Ozair Osman |


