Dr Joanna Achinger-Kawecka
Group Leader, SAiGENCI
SAIGENCI
College of Health
Eligible to supervise Masters and PhD - email supervisor to discuss availability.
Dr Joanna Achinger-Kawecka is a Group Leader at SAiGENCI, University of Adelaide, and an emerging leader in cancer epigenomics and 3D genome biology.She completed her PhD at the University of Tübingen, Germany, supported by the prestigious Marie Curie Fellowship, and undertook postdoctoral training at the Garvan Institute of Medical Research, where she later established her research group. Her pioneering lead-author work has been published in high-impact journals, including Nature Communications, Nature Structural & Molecular Biology and Genome Research. She was subsequently awarded NBCF Mavis Robertson Fellowship and PCFA Young Investigator Award and was appointed as a Group Leader at the Garvan Institute.Joanna has secured over $4 million in competitive funding as a chief investigator (>$3.3M as a CIA, including NHMRC Ideas) and has led multiple successful research programs in collaboration with clinician-researchers, resulting in novel pre-clinical discoveries. In partnership with Arima Genomics (USA), she co-developed a high-resolution Capture Hi-C method, now commercialised and widely adopted by the genomics research community. She is an internationally recognized emerging leader in the field, as evidenced by invitations to present at leading international (EACR 2025, EMBO 2022, Gordon Conference 2018) and national (Lorne Cancer 2025, ACBCC 2025, BMH 2024, ComBio 2022, Lorne Genome 2022, Oz Single Cell 2021) conferencesAt SAiGENCI, she leads the 3D Chromatin Organisation Laboratory, integrating multi-omics, preclinical functional genomics, and translational research to investigate how genome folding and transposable elements drive gene deregulation and therapy resistance, with the goal of informing new therapeutic strategies in rare and hard-to-treat cancers.
The 3D Chromatin Organisation Lab (PI Dr Joanna Achinger) is an experimental biology and bioinformatics laboratory studying the principles of 3D genome folding in cancer. The 3D genome architecture brings together genes and distant regulatory elements to orchestrate gene transcription, and has been implicated in many diseases, including cancer.
Our research focuses on understanding the role of 3D chromatin alterations in driving cancer development, progression and treatment resistance. We use cutting-edge methods (multi-omics, single-cell, genome editing as well as various pre-clinical model systems) to study the interplay of the 3D chromatin, epigenome and transcriptome in cancer.
We use hormone-dependent cancers as a model system for experimental design aimed at increasing our understanding of the cancer biology and to accelerate the development of new therapies.
Current Research Projects:
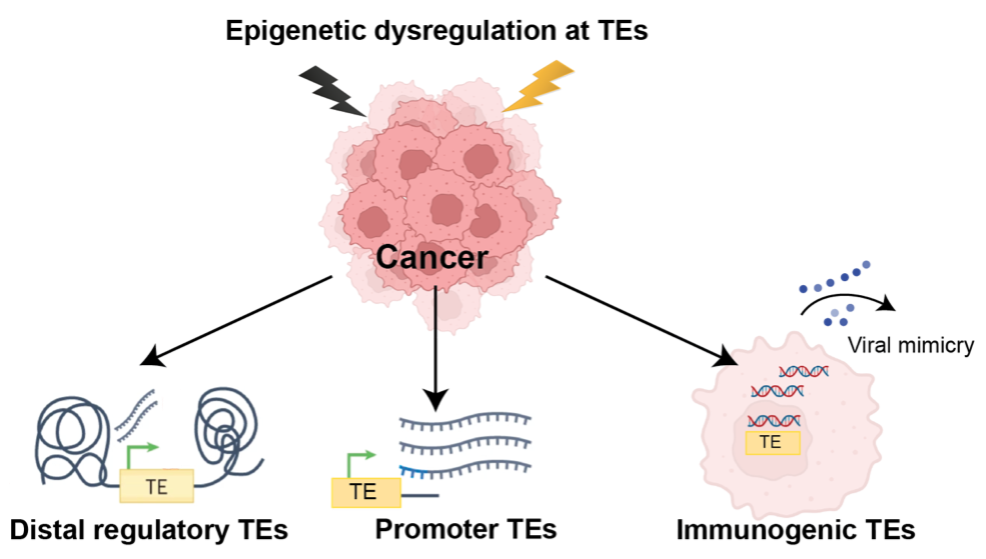
- Exploring the contribution of transposable elements to gene deregulation in cancer
Transposable elements (TEs), known as “jumping genes”, are mobile DNA elements that have expanded within the genome throughout evolution and constitute over 50% of the human genome.
Epigenetic dysregulations are hallmarks of cancer and that provides a particularly fertile ground for TE activation. TEs reactivated in cancer can serve as regulatory elements, including promoters and enhancers, triggering oncogenic transcriptional response. In contrast, induction of TEs through genome-wide epigenetic changes in cancer cells could also potentially trigger anti-tumour immune responses leading to the destruction of cancer cells via “viral mimicry".
The primary objective of this research is to comprehensively delineate the molecular and cellular functions of TEs in the regulatory cancer genome and characterise the epigenetic mechanisms that define their diverse activities. The key aims are to:
- Identify and characterize regulatory TEs across all cancer sub-types in TCGA and determine their transcriptional effects
- Explore how these regulatory TEs contribute to cancer cellular phenotypes using models of TE activation in cancer
- Identify chromatin variants and transcription factors involved in TE regulation and assess their potential for targeting in cancer.
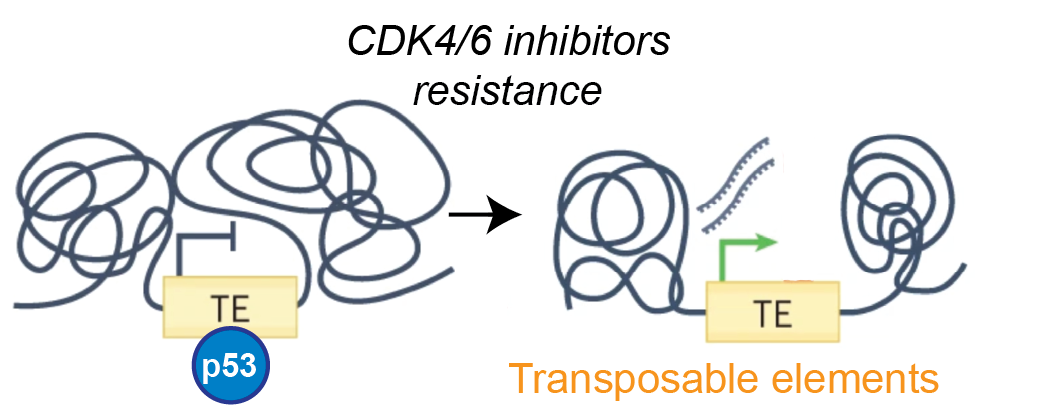
- Targeting transposable elements in CDK4/6i resistant breast cancer
Inhibitors of cyclin-dependant kinases 4 and 6 (CDK4/6i) are now standard of care treatment of metastatic estrogen receptor-positive (ER+) breast cancer. It is expected that all patients with advanced ER+ breast cancer will receive CDK4/6i. However, eventual treatment failure is unfortunately inevitable. No clear second-line treatment strategy exists following the development of CDK4/6i resistance. The widespread emergence of resistance to CDK4/6i is therefore poised to become the major challenge for the management of metastatic ER+ breast cancer.
The TP53 tumour suppressor gene, known as the “guardian of the genome”, is the most frequently altered gene in cancer. It encodes for a transcription factor p53 that triggers a transcriptional program to control cellular stress response. The control of TEs is an important component of p53's function as the guardian of the genome and p53 reactivation is a promising anticancer strategy.
Using in vitro and in vivo pre-clinical models we are uncovering the molecular mechanisms of CDK4/6i resistance and testing if therapeutic targeting of TEs via p53 activation can restore sensitivity and prolong response to CDK4/6 inhibitors in our established cell line and PDX models.
- Elucidating the role of 3D genome structure in lineage plasticity in cancer
Cell lineage plasticity is recognized as a novel mechanism of tumor progression and treatment resistance in cancer. It is mainly driven by dynamic transcriptional and epigenetic changes, however the role of 3D genome structure alterations in driving lineage plasticity is not yet fully understood. Understanding the key 3D genome structure, epigenetic and transcriptional regulators of cancer cell plasticity can yield new avenues for therapy.

- Development and optimzation of new Hi-C technologies
Cellular heterogeneity is a major problem in cancer therapy, as treatment-resistant cells can promote relapse. The advent of single-cell genomics techniques allows the determination of epigenetic and transcriptional profiles of these treatment-resistant populations. By developing new methods to study and integrate the 3D genome, epigenetic and transcriptomics datasets from single-cells, we aim to further understand the molecular mechanisms of treatment resistance and identify new avenues for therapy to target the entire tumour population, or in combination with existing therapies.
| Date | Position | Institution name |
|---|---|---|
| 2025 - ongoing | Head, 3D Chromatin Organisation Laboratory | South Australian ImmunoGENomics Cancer Institute |
| 2025 - ongoing | NBCF Fellow | University of Adelaide |
| 2023 - 2024 | PCFA Young Investigator | Garvan Institute of Medical Research |
| 2021 - 2024 | Head, 3D Epigenome in Cancer Group | Garvan Institute of Medical Research |
| 2021 - 2024 | NBCF Fellow | Garvan Institute of Medical Research |
| 2014 - 2021 | Senior Research Officer | Garvan Institute of Medical Research |
| 2011 - 2014 | Marie Curie ECR Fellow | Dr. Margarete Fischer-Bosch-Institute of Clinical Pharmacology |
| Date | Type | Title | Institution Name | Country | Amount |
|---|---|---|---|---|---|
| 2025 | Fellowship | NBCF Elaine Henry Fellowship | National Breast Cancer Foundation | Australia | - |
| 2023 | Fellowship | PCFA Young Investigator Award | Prostate Cancer Foundation of Australia | Australia | - |
| 2023 | Award | EMBO Travel Award | EMBO | Germany | - |
| 2022 | Award | Franklin Women 2022 Teresa Anderson Award | Franklin Women | Australia | - |
| 2022 | Award | St Vincent’s Campus Rising Star Award | Garvan Institute of Medical Research | Australia | - |
| 2021 | Fellowship | NBCF Mavis Robertson Fellowship | National Breast Cancer Foundation | Australia | - |
| 2019 | Research Award | Estee Lauder Breast Cancer Research Award | Garvan Institute of Medical Research | Australia | - |
| 2019 | Award | EMBO Travel Award | EMBO | Germany | - |
| 2019 | Award | CASS Foundation Award | CASS Foundation | Australia | - |
| 2018 | Award | Ian Potter Foundation Travel Award | Ian Potter Foundation | Australia | - |
| 2017 | Award | Young Garvan Award (finalist) | Garvan Institute of Medical Research | Australia | - |
| 2016 | Award | Heliflite Young Explorer Award | Garvan Institute of Medical Research | Australia | - |
| Date | Institution name | Country | Title |
|---|---|---|---|
| 2011 - 2014 | University of Tubingen | Germany | PhD |
| 2005 - 2011 | Warsaw University of Life Sciences | Poland | MSc |
| Year | Citation |
|---|---|
| 2025 | Tian, L., Jiao, X., Wang, C., Li, D., Ertel, A., Achinger-Kawecka, J., . . . Pestell, R. G. (2025). PPARγ acetylation governs mammary adenocarcinoma tumor growth via acetylated residues that determine DNA sequence-specific binding. Oncogene, 44(37), 3476-3492. |
| 2024 | Achinger-Kawecka, J., Stirzaker, C., Portman, N., Campbell, E., Chia, K. -M., Du, Q., . . . Clark, S. J. (2024). The potential of epigenetic therapy to target the 3D epigenome in endocrine-resistant breast cancer. Nature Structural and Molecular Biology, 31(3), 498-512. Scopus20 WoS19 Europe PMC23 |
| 2024 | Peters, T. J., Meyer, B., Ryan, L., Achinger-Kawecka, J., Song, J., Campbell, E. M., . . . Pidsley, R. (2024). Characterisation and reproducibility of the HumanMethylationEPIC v2.0 BeadChip for DNA methylation profiling. BMC Genomics, 25(1), 251-1-251-23. Scopus27 WoS26 Europe PMC34 |
| 2024 | Chen, W., Zeng, Y. C., Achinger-Kawecka, J., Campbell, E., Jones, A. K., Stewart, A. G., . . . Clark, S. J. (2024). Machine learning enables pan-cancer identification of mutational hotspots at persistent CTCF binding sites. Nucleic Acids Research, 52(14), 8086-8099. Scopus5 WoS5 Europe PMC5 |
| 2023 | Achinger-Kawecka, J., Correa, S., Hu, J., Li, G., Lindeboom, R. G. H., Misale, S., . . . Watson, C. J. (2023). The 2023 generation. Nature Cancer, 4(12), 1630-1635. Europe PMC1 |
| 2023 | Hastings, J. F., Latham, S. L., Kamili, A., Wheatley, M. S., Han, J. Z. R., Wong-Erasmus, M., . . . Croucher, D. R. (2023). Memory of stochastic single-cell apoptotic signaling promotes chemoresistance in neuroblastoma. Science Advances, 9(9), eabp8314-1-eabp8314-23. Scopus15 WoS14 Europe PMC12 |
| 2022 | Brown, L. J., Achinger-Kawecka, J., Portman, N., Clark, S., Stirzaker, C., & Lim, E. (2022). Epigenetic Therapies and Biomarkers in Breast Cancer. Cancers, 14(3), 20 pages. Scopus43 WoS36 Europe PMC33 |
| 2021 | Du, Q., Smith, G. C., Luu, P. L., Ferguson, J. M., Armstrong, N. J., Caldon, C. E., . . . Clark, S. J. (2021). DNA methylation is required to maintain both DNA replication timing precision and 3D genome organization integrity.. Cell reports, 36(12), 109722. Scopus42 WoS44 Europe PMC51 |
| 2021 | Giles, K. A., Gould, C. M., Achinger-Kawecka, J., Page, S. G., Kafer, G. R., Rogers, S., . . . Taberlay, P. C. (2021). BRG1 knockdown inhibits proliferation through multiple cellular pathways in prostate cancer.. Clinical epigenetics, 13(1), 37. Scopus20 WoS19 Europe PMC17 |
| 2020 | Achinger-Kawecka, J., Valdes-Mora, F., Luu, P. -L., Giles, K. A., Caldon, C. E., Qu, W., . . . Clark, S. J. (2020). Epigenetic reprogramming at estrogen-receptor binding sites alters 3D chromatin landscape in endocrine-resistant breast cancer.. Nature communications, 11(1), 320. Scopus108 WoS108 Europe PMC125 |
| 2020 | Khoury, A., Achinger-Kawecka, J., Bert, S. A., Smith, G. C., French, H. J., Luu, P. -L., . . . Clark, S. J. (2020). Constitutively bound CTCF sites maintain 3D chromatin architecture and long-range epigenetically regulated domains.. Nature communications, 11(1), 54. Scopus64 WoS61 Europe PMC75 |
| 2019 | Giles, K. A., Gould, C. M., Du, Q., Skvortsova, K., Song, J. Z., Maddugoda, M. P., . . . Taberlay, P. C. (2019). Integrated epigenomic analysis stratifies chromatin remodellers into distinct functional groups. Epigenetics and Chromatin, 12(1), 19 pages. Scopus22 WoS22 Europe PMC24 |
| 2018 | Fuksiewicz, M., Kotowicz, B., Rutkowski, A., Achinger-Kawecka, J., Wagrodzki, M., & Kowalska, M. M. (2018). The assessment of clinical usage and prognostic value of YKL-40 serum levels in patients with rectal cancer without distant metastasis. Technology in Cancer Research and Treatment, 17, 8 pages. Scopus17 WoS16 Europe PMC13 |
| 2017 | Achinger-Kawecka, J., & Clark, S. J. (2017). Disruption of the 3D cancer genome blueprint. Epigenomics, 9(1), 47-55. Scopus36 WoS36 Europe PMC35 |
| 2016 | Achinger-Kawecka, J., Taberlay, P. C., & Clark, S. J. (2016). Alterations in three-dimensional organization of the cancer genome and epigenome. Cold Spring Harbor Symposia on Quantitative Biology, 81(1), 41-51. Scopus27 WoS24 Europe PMC24 |
| 2016 | Taberlay, P. C., Achinger-Kawecka, J., Lun, A. T. L., Buske, F. A., Sabir, K., Gould, C. M., . . . Clark, S. J. (2016). Three-dimensional disorganization of the cancer genome occurs coincident with long-range genetic and epigenetic alterations. Genome Research, 26(6), 719-731. Scopus236 WoS223 Europe PMC251 |
| 2013 | Hoppe, R., Achinger-Kawecka, J., Winter, S., Fritz, P., Lo, W. Y., Schroth, W., & Brauch, H. (2013). Increased expression of miR-126 and miR-10a predict prolonged relapse-free time of primary oestrogen receptor-positive breast cancer following tamoxifen treatment. European Journal of Cancer, 49(17), 3598-3608. Scopus73 WoS73 Europe PMC70 |
| 2013 | Hoppe, R., Achinger-Kawecka, J., Winter, S., Fritz, P., Lo, W. -Y., Schroth, W., & Brauch, H. (2013). Abstract 1938: Increased miR-126 and miR-375 expression in primary ER-positive breast cancer predict longer relapse-free time following treatment with tamoxifen.. Cancer Research, 73(8_Supplement), 1938. |
| - | Stirzaker, C., Chia, K. M., Portman, N., Milioli, H. H., Clifton, S., Achinger-Kawecka, J., . . . Clark, S. J. (2019). DNA demethylation agents as a therapeutic approach in endocrine-resistant breast cancer. Oncology Abstracts. |
| Year | Citation |
|---|---|
| 2024 | Tian, L., Jiao, X., Wang, C., Ertel, A., Soccio, R., Chen, E. R., . . . Pestell, R. G. (2024). PPAR gamma acetylation governs mammary adenocarcinoma tumor growth via acetylated residues that determine DNA sequence-specific binding. In CANCER RESEARCH Vol. 84 (pp. 2 pages). CA, San Diego: AMER ASSOC CANCER RESEARCH. DOI |
| Year | Citation |
|---|---|
| 2024 | Campbell, E., Laven-Law, G., Smith, G., Peters, T., Colino-Sanguino, Y., Moulder, D., . . . Achinger-Kawecka, J. (2024). Androgen stimulation rapidly reorganizes temporal 3D genome and epigenome states to trigger AR-mediated transcription in prostate cancer. DOI |
| 2021 | Achinger-Kawecka, J., Stirzaker, C., Portman, N., Campbell, E., Chia, K. -M., Du, Q., . . . Clark, S. (2021). Epigenetic therapy targets the 3D epigenome in endocrine-resistant breast cancer. DOI |
| 2020 | Giles, K. A., Gould, C. M., Achinger-Kawecka, J., Page, S. G., Kafer, G., Luu, P. -L., . . . Taberlay, P. C. (2020). BRG1 promotes transcriptional patterns that are permissive to proliferation in cancer cells. DOI |
| 2020 | Du, Q., Smith, G. C., Luu, P. L., Ferguson, J. M., Armstrong, N. J., Caldon, C. E., . . . Clark, S. J. (2020). DNA methylation is required to maintain DNA replication timing precision and 3D genome integrity. DOI |
2026 - 2030 The Sylvia and Charles Viertel Charitable Foundation - Senior Medical Research Fellowship
2025 - 2029 National Breast Cancer Foundation (NBCF) Elaine Henry Fellowship ($20k)
2025 - 2029 National Breast Cancer Foundation (NBCF) Research Project Grant (PI, $1.098M): Directing p53 to transposable elements to promote antitumour immunity in CDK4/6 inhibitors resistant breast cancer.
2025 - 2026 National Computing Infrastructure (NCI) NCMAS Grant (PI): Establishing the regulatory role of transposable elements in cancer.
2024 - 2026 NHMRC Ideas Grant (CIA, $1.09M): Assessing the role of the tumour ecosystem in epigenetic therapy response in endocrine resistant breast cancer.
2024 - 2026 Cancer Council NSW (CID, $450k): Resolving Prostate Cancer: Elucidating the epigenetics of tumour microenvironment cells to advance understanding of disease aetiology and improve patient diagnosis.
2024 - 2025 Prostate Cancer Foundation of Australia (PCFA) Young Investigator Award (PI, $100K): Targeting epigenetic hallmarks in neuroendocrine-like prostate cancer.
2023 - 2024 UNSW Cancer Theme EMCR Seed Grant (PI, $50k): Targeting enhancers to overcome breast cancer resistance to CDK4/6 inhibition.
2021 - 2024 National Breast Cancer Foundation (NBCF) Investigator Initiated Research Scheme and NBCF Mavis Robertson Fellowship (PI, $366k): 3D Epigenome as a Target for Epigenetic Therapies in Endocrine-Resistant Breast Cancer.
2020 - 2023 Cancer Council NSW Project Grant (CIA, $450k): Using Epigenetic Therapies to Overcome Endocrine Resistance in Breast Cancer.
2020 - 2021 UNSW Cellular Genomics Futures Institute Seed Grant (PI, $100k): Decoding 3D genome architecture in individual single-cells to establish molecular mechanisms of gene regulation in cancer.
2017 - 2018 National Breast Cancer Foundation (NBCF) Innovator Grant (Co-I, $192k): Genetic perturbations to the 3D genome architecture: Implications for endocrine resistance in breast cancer.
Teaching, Mentoring and Supervision:
Postdoctoral Fellows (Present):
Dr Daniel Thomson
Dr Fiona Zhou
Dr Dayna Challis
Geraldine Laven Law (Research Manager)
Postdoctoral Fellows (Past):
Dr Qian Du (Now postdoc at University of Copenhagen)
Dr Kate Giles (Now postdoc at CMRI, Sydney)
Teaching:
2022 - 2024 Co-ordinator of TKCC Cancer Seminars, Garvan Institute
2022 - Guest Lecturer, School of Biotechnology and Biomolecular Sciences, UNSW
2022 - Guest Lecturer, St George and Sutherland Clinical School Research in Progress Meetings, University of Sydney
2019 - Guest Lecturer, Brain Sciences UNSW Colloquia: “Genetic and Epigenetic Contributions”, Black Dog Institute
| Date | Role | Research Topic | Location | Program | Supervision Type | Student Load | Student Name |
|---|---|---|---|---|---|---|---|
| 2023 - ongoing | Principal Supervisor | Targeting epigenetic hallmarks in advanced prostate cancer | UNSW Sydney | Doctor of Philosophy | Doctorate | Full Time | Anthony Rodrigues |
| 2022 - ongoing | Principal Supervisor | Targeting the 3D epigenome to overcome treatment resistance in cancer | UNSW Sydney | Doctor of Philosophy | Doctorate | Full Time | Elyssa Campbell |
| 2021 - ongoing | Co-Supervisor | Chromatin architecture and epigenomic regulation of cardiomyocyte regeneration | UNSW Sydney | Doctor of Philosophy | Doctorate | Full Time | Gabrielle Smith |
| 2015 - 2019 | Co-Supervisor | BRG1 ATP-Dependent Chromatin Remodelling in Prostate Cancer: Epigenetic Consequences | UNSW Sydney | Doctor of Philosophy | Doctorate | Full Time | Kate Giles |
| Date | Role | Committee | Institution | Country |
|---|---|---|---|---|
| 2025 - ongoing | Co-Chair | High Performance Computing Committee | SAiGENCI | Australia |
| 2025 - ongoing | Member | Australian Bioinformatics and Computational Biology Society | ABACBS | Australia |
| 2024 - ongoing | Member | Chair and Programme Co-organizer, Biomolecular Horizons 2024 | ComBio | Australia |
| 2024 - 2024 | Member | UNSW Cancer Theme Strategic Advisory Committee | UNSW | Australia |
| 2024 - ongoing | Member | Australian Academy of Science, Science at the Shine Dome EMCR Committee | Australian Academy of Science | Australia |
| 2021 - 2024 | Member | Higher Research Degree Committee | Garvan Institute of Medical Research | Australia |
| 2020 - 2023 | Member | Garvan Engagement Committee | Garvan Institute of Medical Research | Australia |
| 2017 - 2018 | Co-Chair | ASMR NSW Annual Scientific Meeting Committee | ASMR | Australia |
| 2015 - 2016 | Co-Chair | Garvan Postdoctoral Developmental Committee | Garvan Institute of Medical Research | Australia |
| Date | Role | Membership | Country |
|---|---|---|---|
| 2024 - ongoing | Member | Australian Bioinformatics and Computational Biology Society | Australia |
| 2014 - ongoing | Member | Australian Society for Medical Research | Australia |
| 2014 - ongoing | Member | Australasian Epigenetics Alliance | Australia |
| 2011 - ongoing | Member | American Association for Cancer Research | United States |
| Date | Title | Engagement Type | Institution | Country |
|---|---|---|---|---|
| 2024 - ongoing | Researcher Showcase (video) | Public Community Engagement | Cancer Council NSW and Box Rallies | - |
| 2024 - 2024 | New hope for breast cancer patients | Public Community Engagement | The Australian (newspaper) | - |
| 2023 - 2023 | Interview, NBCF 30th Anniversary | Public Community Engagement | National Breast Cancer Foundation | - |
| 2020 - ongoing | Researcher Showcase Discussion Panel (Public Event) | Public Community Engagement | Garvan Institute of Medical Research | - |
| Date | Title | Type | Institution | Country |
|---|---|---|---|---|
| 2025 - ongoing | DFG - Deutsche Forschungsgemeinschaft | Grant Assessment | DFG | Germany |
| 2025 - ongoing | Worldwide Cancer Research Foundation | Peer Review | Worldwide Cancer Research Foundation | - |
| 2024 - ongoing | Fondation pour la Recherche Médicale (France) | Grant Assessment | Fondation pour la Recherche Médicale (France) | France |
| 2024 - ongoing | Neurological Foundation of New Zealand | Grant Assessment | Neurological Foundation of New Zealand | New Zealand |
| 2024 - ongoing | Diabetes Australia | Grant Assessment | Diabetes Australia | Australia |
| 2023 - ongoing | Garvan-Estée Lauder Award | Grant Assessment | Garvan Institute of Medical Research | - |
| 2023 - ongoing | Young TAD Award | Peer Review | Genome Organisation Australia | - |
| 2023 - ongoing | Harry Perkins Institute of Medical Research | Grant Assessment | Harry Perkins Institute of Medical Research | Australia |
| 2021 - ongoing | NHMRC Ideas Grants | Peer Review | NHMRC | - |
| 2020 - ongoing | European Research Council MSCA Horizon | Grant Assessment | European Research Council | - |
| 2020 - ongoing | NCN Poland | Grant Assessment | NCN Poland | Poland |







Available For Media Comment.