
APrf Iain Searle
Associate Professor
School of Biological Sciences
College of Sciences
Eligible to supervise Masters and PhD - email supervisor to discuss availability.
Epigenetic and post-transcriptional gene regulation
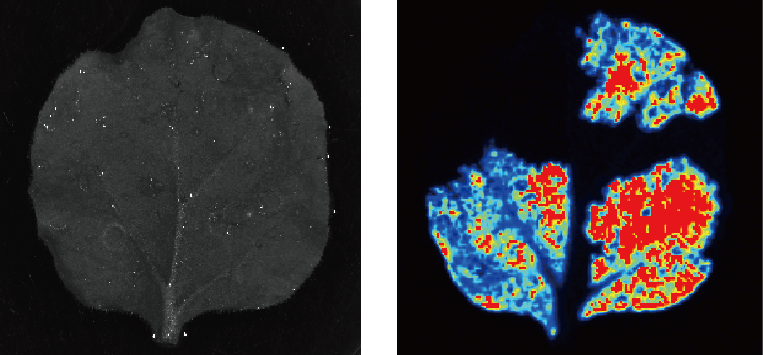
Our research in plant and animal epigenetics and RNA modifications has been driven by a strong interest in understanding and elucidating the molecular mechanisms of how plants and animals adapt to environmental signalling and how speciation barriers evolve in plants. We use a combination of next-generation sequencing, bioinformatics and synthetic biology to describe and elucidate the function of these epigenetic components.
Epigenetics, broadly defined, refers to all genetic information not encoded in the DNA sequence, with the best-understood consequence of epigenetic modifications being the regulation of gene expression. Although gene transcription and protein synthesis is required for plant development, the full repertoire of epigenetic mechanisms underpinning biological adaptation and speciation barriers remain equivocal.
The majority of the genome is pervasively transcribed such that more than 90% of transcriptional activity is related to the production of various long non-coding RNAs (lncRNA), which possess no or limited protein-coding capacity. Expansion of these transcriptionally active non-coding sequences in plant and animal genomes appears to have occurred primarily in higher multicellular organisms. We have identified thousands of novel lncRNAs transcribed in plants.
Furthermore, we have demonstrated RNA modifications, for example 5-methylcytosine (m5C) play an important role in extending the transcriptional space. Thus, both RNA modifications and lncRNAs represent attractive candidates for achieving context- and stimulus-specific epigenetic regulation of gene expression.
Honours, Masters, PhD and postdoctoral synthetic biology and biotechnology projects in epigenetics and bioinformatics are available. Email me at, iain.searle@adelaide.edu.au, for further information.
Developing sustainable biocontrols for agricultural and horticultural diseases

20% to 40% of global crop production is lost to pests, and plant diseases cost the global economy over $200 billion annually. Crown gall disease caused by Agrobacterium and related spp. like Allorhizobium can significantly reduce the yield of infected trees, examples include almonds and walnuts, and young grapevines.
Crown gall disease in young grapevines is a global problem in nurseries and vineyards. Symptoms include gall formation at graft and wound sites, stunted growth and in later stages of the disease progression decline in vine vigour and yield. As there is currently no cure, we are developing the next generation of sustainable, environmentally friendly, biocontrols with cutting-edge technologies like CRISPR and third-generation sequencing.
There are a range of biochemistry and synthetic biology projects available.
Honours, Masters, PhD and postdoctoral CRISPR and biotechnology projects in are available. Email me at, iain.searle@adelaide.edu.au, for further information.
Relevant papers from our group characterising grapevine crown gall pathogenic and non-pathogenic strains
(1) Genome sequence of Grapevine Crown Gall pathogenic strain A. vitis K306
(2) Genome sequence of Grapevine Crown Gall pathogenic strain A. vitis K377
(3) Genome sequence of nonpathogenic A. vitis F2/5
Relevant peer-reviewed research from our collaborators
(1) PhD thesis on Biological control of Crown Gall Disease in Australian Grape vine nurseries (2001)- of Dr. Alex Keegan. Former student of collaborator Prof. Allen Kerr
(2) Detection of pathogenic A. vitis in grapevines (2016)- Collaborator Prof. Thomas Burr
(3) Biochemical characterisation of crown gall compounds (1984)- Collaborator Dr. Maarten Ryder
Sustainable crops for arid Australia

Australia is a hot, dry country and often water limits crop yields. We are developing Common Vetch, Vicia sativa, as a high protein, drought tolerant legume for arid Australia. We are accelerating common vetch improvement by; developing extensive genomic resources using a combination of second and third generation sequencing and bioinformatics, combining these large datasets with phenotyping information, and then using next generation breeding technologies (NBT), like CRISPR genome editing. We are working closely with the National Vetch Breeding program at SARDI (Waite campus) to further develop vetch.
Honours, Masters, PhD and postdoctoral CRISPR and biotechnology projects in are available. Email me at, iain.searle@adelaide.edu.au, for further information.
Pollination Mechanisms

We are interested in how plants promote outcrossing to increase population fitness. We are using the model crop Brassica rapa to solve the molecular basis of an outcrossing mechanism. Conversely, these outcrossing mechanisms can decrease seed and crop yields and we are working with industrial partners to determine if we can increase crops yields.
Honours, Masters, PhD and postdoctoral ecology and industry projects in are available. Email me at, iain.searle@adelaide.edu.au, for further information.
Interested in joining our laboratory as an undergraduate placement student or postgraduate student? What to join our mentor programme? Ready to excel and be challenged with fascinating biological questions? Please don't hesitate to email me- iain.searle@adelaide.edu.au.
Publications
Our group's publications are available here.
Contact
| Date | Position | Institution name |
|---|---|---|
| 2018 - ongoing | Senior Lecturer | University of Adelaide |
| 2014 - 2018 | ARC Future Fellow | University of Adelaide |
| 2009 - 2014 | ARC QEII Fellow | Australian National University |
| Language | Competency |
|---|---|
| German | Can speak and understand spoken |
| Date | Institution name | Country | Title |
|---|---|---|---|
| 2004 | University of Queensland | Australia | PhD |
| 1996 | University of Queensland | Australia | B Ag Sci Hons |
| Date | Title | Institution | Country |
|---|---|---|---|
| 2007 - 2009 | Wellcome Trust VIP Fellow | University of Cambridge | United Kingdom |
| 2006 - 2007 | Marie Curie Postdoctoral Fellow | The Sainsbury Laboratory | United Kingdom |
| 2002 - 2006 | Max Planck Postdoctoral Fellow | Max Planck Institute for Plant Breeding Research | Germany |
| Year | Citation |
|---|---|
| 2025 | Zhao, L., Ma, X., Ma, W., Wang, Z., Luo, D., Zhou, Q., . . . Liu, Z. (2025). MsPYL6 and MsPYL9 improves drought tolerance by regulating stomata in alfalfa (Medicago sativa). The Plant Journal, 122(6), e70265-1-e70265-16. Scopus5 WoS6 Europe PMC3 |
| 2025 | Liu, W., Zhao, X., Li, Y., Zhou, Q., Searle, I. R., Xie, W., . . . Liu, Z. (2025). Genome-Wide Association Study Reveals the Genetic Architecture and Key Drought and Yield Related Genes in Common Vetch (Vicia sativa L.). Plant Biotechnology Journal, 1-16. |
| 2025 | Riley, S., Nguyen, V., Bhattacharjee, R., Ng, P. Q., Ritchie, T., Kamath, K. S., . . . Searle, I. R. (2025). TMT-based quantitative proteomic assessment of Vicia sativa induced neurotoxicity by β-cyano-L-alanine and γ-glutamyl-β-cyano-L-alanine in SH-SY5Y cells.. Scientific reports. |
| 2024 | Li, M., Pu, J., Jia, C., Luo, D., Zhou, Q., Fang, X., . . . Liu, Z. (2024). The genome of Vicia sativa ssp. amphicarpa provides insights into the role of terpenoids in antimicrobial resistance within subterranean fruits. Plant Journal, 119(6), 2654-2671. Scopus2 WoS2 Europe PMC1 |
| 2024 | Xue, F., Zhang, J., Wu, D., Sun, S., Fu, M., Wang, J., . . . Liang, W. (2024). m<sup>6</sup>A demethylase OsALKBH5 is required for double-strand break formation and repair by affecting mRNA stability in rice meiosis.. The New phytologist, 244(6), 2326-2342. Scopus7 WoS7 Europe PMC6 |
| 2023 | Chmielowska-Bąk, J., Searle, I. R., Wakai, T. N., & Arasimowicz-Jelonek, M. (2023). The role of epigenetic and epitranscriptomic modifications in plants exposed to non-essential metals. Frontiers in Plant Science, 14, 1278185-1-1278185-14. Scopus7 WoS6 Europe PMC6 |
| 2023 | Nguyen, V., & Searle, I. (2023). Keeping it cool.. eLife, 12, e86885. Scopus1 WoS1 Europe PMC1 |
| 2023 | Safdar, L. B., Dugina, K., Saeidan, A., Yoshicawa, G. V., Caporaso, N., Gapare, B., . . . Fisk, I. D. (2023). Reviving grain quality in wheat through non‐destructive phenotyping techniques like hyperspectral imaging. Food and Energy Security, 12(5), 1-21. Scopus13 WoS12 Europe PMC4 |
| 2023 | Safdar, L. B., Foulkes, M. J., Kleiner, F. H., Searle, I. R., Bhosale, R. A., Fisk, I. D., & Boden, S. A. (2023). Challenges facing sustainable protein production: Opportunities for cereals. Plant Communications, 4(6), 100716-1-100716-15. Scopus52 WoS41 Europe PMC20 |
| 2022 | Xi, H., Grist, J., Ryder, M., & Searle, I. (2022). Complete Genome Sequence Data of the Grapevine Crown Gall inhibiting bacteria Allorhizobium vitis Strain F2/5.. Mol Plant Microbe Interact, 35(2), 174-176. Scopus2 |
| 2022 | Nguyen, V., & Searle, I. R. (2022). An efficient root transformation system for recalcitrant Vicia sativa. Frontiers in Plant Science, 12, 781014-1-781014-11. Scopus4 WoS4 Europe PMC2 |
| 2022 | Xi, H., Nguyen, V., Ward, C., Liu, Z., & Searle, I. R. (2022). Chromosome-level assembly of the common vetch (Vicia sativa) reference genome. Gigabyte, 2022, 1-19. Scopus22 Europe PMC17 |
| 2022 | Li, M., Zhao, L., Zhou, Q., Fang, L., Luo, D., Liu, W., . . . Liu, Z. (2022). Transcriptome and Coexpression Network Analyses Provide In-Sights into the Molecular Mechanisms of Hydrogen Cyanide Synthesis during Seed Development in Common Vetch (Vicia sativa L.). International Journal of Molecular Sciences, 23(4), 2275-1-2275-15. Scopus7 WoS7 Europe PMC3 |
| 2022 | Dixon, L. E., Pasquariello, M., Badgami, R., Levin, K. A., Poschet, G., Ng, P. Q., . . . Boden, S. A. (2022). MicroRNA-resistant alleles of HOMEOBOX DOMAIN-2 modify inflorescence branching and increase grain protein content of wheat. Science Advances, 8(19), 1-16. Scopus31 WoS30 Europe PMC22 |
| 2022 | Riley, S., Lale, A., Nguyen, V., Xi, H., Wilkinson, K., Searle, I. R., & Fisk, I. (2022). Volatile profiles of commercial vetch prepared via different processing methods. Food Chemistry, 395, 15 pages. Scopus7 WoS7 Europe PMC3 |
| 2021 | David, R., Ng, P. Q., Smith, L. M., & Searle, I. R. (2021). Novel allele <i>elh</i> of the <i>UBP14</i> gene affects plant organ size via cell expansion in <i>Arabidopsis thaliana.</i>. microPublication biology, 2021. Europe PMC2 |
| 2021 | Xi, H., Ryder, M., & Searle, I. R. (2021). Near-Complete Genome Assembly of the Grapevine Crown Gall Pathogen Allorhizobium vitis Strain K377. Microbiology Resource Announcements, 10(39), 1-2. Scopus2 |
| 2021 | Xi, H., Nguyen, V., Ward, C., Liu, Z., & Searle, I. (2021). Chromosome-level assembly of the common vetch reference genome (Vicia sativa). GIGABYTE, 2022, 19 pages. WoS3 Europe PMC1 |
| 2021 | Lim, S. M., Choo, J. M., Li, H., O'Rielly, R., Carragher, J., Rogers, G. B., . . . Muhlhausler, B. (2021). A high amylose wheat diet improves gastrointestinal health parameters and gut microbiota in male and female mice. Foods, 10(2), 1-18. Scopus12 WoS12 Europe PMC8 |
| 2021 | Jia, C., Dong, D., Zhou, Q., Searle, I. R., & Liu, Z. (2021). Significant cell difference of pod ventral suture in shatter‐resistant and shatter‐susceptible common vetch accessions. Crop Science, 61(3), 1-11. Scopus8 WoS8 |
| 2020 | Sinha, P., Singh, V. K., Saxena, R. K., Kale, S. M., Li, Y., Garg, V., . . . Varshney, R. K. (2020). Genome-wide analysis of epigenetic and transcriptional changes associated with heterosis in pigeonpea.. Plant biotechnology journal, 18(8), 1697-1710. Scopus54 WoS49 Europe PMC36 |
| 2020 | Nguyen, V., & Searle, I. R. (2020). An efficient root transformation system for recalcitrant<i>Vicia sativa</i>. |
| 2020 | Xi, H., Ryder, M., & Searle, I. (2020). Complete genome sequence of Allorhizobium vitis strain K306, the causal agent of grapevine crown gall. Microbiologial Resource Annoucements, 9(29), e00565-20-1-e00565-20-3. Scopus3 WoS3 Europe PMC2 |
| 2020 | Nguyen, V., Riley, S., Nagel, S., Fisk, I., & Searle, I. R. (2020). Common vetch: a drought tolerant, high protein neglected leguminous crop with potential as a sustainable food source. Frontiers in Plant Science, 11, 818-1-818-7. Scopus39 WoS36 Europe PMC22 |
| 2020 | Xiao, L., Jiang, S., Huang, P., Chen, F., Wang, X., Cheng, Z., . . . Zhang, X. -M. (2020). Two Nucleoporin98 homologous genes jointly participate in the regulation of starch degradation to repress senescence in Arabidopsis. BMC Plant Biology, 20(1), 292-1-292-15. Scopus14 WoS17 Europe PMC16 |
| 2019 | Lim, S. M., Page, A. J., Li, H., Carragher, J., Searle, I., Robertson, S., & Muhlhausler, B. (2019). Sexually Dimorphic Response of Increasing Dietary Intake of High Amylose Wheat on Metabolic and Reproductive Outcomes in Male and Female Mice.. Nutrients, 12(1), 14 pages. Scopus6 WoS4 Europe PMC3 |
| 2019 | Guo, Q., Ng, P. Q., Shi, S., Fan, D., Li, J., Zhao, J., . . . Searle, I. R. (2019). Arabidopsis TRM5 encodes a nuclear-localised bifunctional tRNA guanine and inosine-N1-methyltransferase that is important for growth. PLoS ONE, 14(11), 1-26. Scopus23 WoS22 Europe PMC21 |
| 2019 | David, R., Kortschak, R. D., & Searle, I. R. (2019). The root hair defective phenotype of Arabidopsis thaliana Pol IV subunit mutant nrpd1a-3 is associated with a deletion in RHD6. microPublication Biology, 2019, 1-4. Europe PMC3 |
| 2018 | Wang, D., Gu, J., David, R., Wang, Z., Yang, S., Searle, I., . . . Timmis, J. (2018). Experimental reconstruction of double-stranded break repair-mediated plastid DNA insertion into the tobacco nucleus. The Plant Journal, 93(2), 227-234. Scopus8 WoS8 Europe PMC9 |
| 2017 | Crisp, P., Ganguly, D., Smith, A., Murray, K., Estavillo, G., Searle, I., . . . Pogson, B. (2017). Rapid recovery gene downregulation during excess-light stress and recovery in Arabidopsis. The Plant Cell, 29(8), 1836-1863. Scopus79 WoS81 Europe PMC81 |
| 2017 | Liu, L., Jiang, Y., Zhang, X., Wang, X., Wang, Y., Han, Y., . . . Chen, F. (2017). Two SUMO proteases SUMO PROTEASE RELATED TO FERTILITY1 and 2 are required for fertility in arabidopsis. Plant Physiology, 175(4), 1703-1719. Scopus35 WoS34 Europe PMC30 |
| 2017 | David, R., Burgess, A., Parker, B., Li, J., Pulsford, K., Sibbritt, T., . . . Searle, I. (2017). Transcriptome-wide Mapping of RNA 5-Methylcytosine in Arabidopsis mRNAs and non-coding RNAs. The Plant cell, 29(3), 16.00751-1-16.0075-30. Scopus207 WoS197 Europe PMC191 |
| 2017 | Wang, D., Qu, Z., Yang, L., Zhang, Q., Liu, Z., Do, T., . . . Zhu, J. (2017). Transposable elements (TEs) contribute to stress-related long intergenic noncoding RNAs in plants. Plant Journal, 90(1), 133-146. Scopus130 WoS116 Europe PMC97 |
| 2016 | Ashby, R., Forêt, S., Searle, I., & Maleszka, R. (2016). MicroRNAs in Honey Bee Caste Determination. Scientific Reports, 6(1), 15 pages. Scopus118 WoS104 Europe PMC76 |
| 2016 | Burgess, A., David, R., & Searle, I. (2016). Deciphering the epitranscriptome: a green perspective. Journal of Integrative Plant Biology, 58(10), 822-835. Scopus32 WoS32 Europe PMC28 |
| 2016 | Chen, M., Xu, J., Devis, D., Shi, J., Ren, K., Searle, I., & Zhang, D. (2016). Origin and functional prediction of pollen allergens in plants. Plant Physiology, 172(1), 341-357. Scopus31 WoS29 Europe PMC19 |
| 2015 | Burgess, A., David, R., & Searle, I. (2015). Conservation of tRNA and rRNA 5-methylcytosine in the kingdom Plantae. BMC Plant Biology, 15(1), 199-1-199-17. Scopus98 WoS101 Europe PMC90 |
| 2015 | Liu, H., Searle, I., Mather, D., Able, A., & Able, J. (2015). Morphological, physiological and yield responses of durum wheat to pre-anthesis water-deficit stress are genotype-dependent. Crop and Pasture Science, 66(10), 1024-1038. Scopus75 WoS61 |
| 2015 | Liu, H., Searle, I., Watson-Haigh, N., Baumann, U., Mather, D., Able, A., & Able, J. (2015). Genome-wide identification of microRNAs in leaves and the developing head of four durum genotypes during water deficit stress. PLoS One, 10(11), e0142799-1-e0142799-30. Scopus50 WoS44 Europe PMC32 |
| 2013 | Cazzonelli, C., Vanstraelen, M., Simon, S., Yin, K., Carron-Arthur, A., Nisar, N., . . . Pogson, B. (2013). Role of the Arabidopsis PIN6 auxin transporter in auxin homeostasis and auxin-mediated development. PLoS One, 8(7), 1-14. Scopus75 WoS72 Europe PMC52 |
| 2012 | Campoli, C., Drosse, B., Searle, I., Coupland, G., & von Korff, M. (2012). Functional characterisation of HvCO1, the barley (Hordeum vulgare) flowering time ortholog of CONSTANS. The Plant Journal, 69(5), 868-880. Scopus140 WoS131 Europe PMC111 |
| 2011 | Gursanscky, N. R., Searle, I. R., & Carroll, B. J. (2011). Mobile microRNAs hit the target. Traffic, 12(11), 1475-1482. Scopus12 WoS11 Europe PMC9 |
| 2011 | Burgess, A., & Searle, I. R. (2011). The clock primes defense at dawn. Immunology and Cell Biology, 89(6), 661-662. Scopus4 WoS4 Europe PMC3 |
| 2011 | Indrasumunar, A., Searle, I., Lin, M., Kereszt, A., Men, A., Carroll, B., & Gresshoff, P. (2011). Nodulation factor receptor kinase 1α controls nodule organ number in soybean (Glycine max L. Merr). The Plant Journal, 65(1), 39-50. Scopus124 WoS117 Europe PMC101 |
| 2011 | Castro Marín, I., Loef, I., Bartetzko, L., Searle, I., Coupland, G., Stitt, M., & Osuna, D. (2011). Nitrate regulates floral induction in Arabidopsis, acting independently of light, gibberellin and autonomous pathways. Planta, 233(3), 539-552. Scopus168 WoS157 Europe PMC108 |
| 2010 | Searle, I. R., Pontes, O., Melnyk, C. W., Smith, L. M., & Baulcombe, D. C. (2010). JMJ14, a JmjC domain protein, is required for RNA silencing and cell-to-cell movement of an RNA silencing signal in Arabidopsis. Genes and Development, 24(10), 986-991. Scopus101 WoS97 Europe PMC93 |
| 2010 | Indrasumunar, A., Kereszt, A., Searle, I., Miyagi, M., Li, D., Nguyen, C. D. T., . . . Gresshoff, P. M. (2010). Inactivation of duplicated nod factor receptor 5 (NFR5) genes in recessive loss-of-function non-nodulation mutants of allotetraploid soybean (Glycine max L. Merr.). Plant and Cell Physiology, 51(2), 201-214. Scopus112 WoS107 Europe PMC88 |
| 2009 | Huang, L., Jones, A. M. E., Searle, I., Patel, K., Vogler, H., Hubner, N. C., & Baulcombe, D. C. (2009). An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nature Structural and Molecular Biology, 16(1), 91-93. Scopus116 WoS104 Europe PMC98 |
| 2007 | Corbesier, L., Vincent, C., Jang, S., Fornara, F., Fan, Q., Searle, I., . . . Coupland, G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science, 316(5827), 1030-1033. Scopus1873 WoS1772 Europe PMC1471 |
| 2007 | Smith, L. M., Pontes, O., Searle, I., Yelina, N., Yousafzai, F. K., Herr, A. J., . . . Baulcombe, D. C. (2007). An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell, 19(5), 1507-1521. Scopus246 WoS228 Europe PMC214 |
| 2006 | Searle, I., He, Y., Turck, F., Vincent, C., Fornara, F., Kröber, S., . . . Coupland, G. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes and Development, 20(7), 898-912. Scopus757 WoS717 Europe PMC610 |
| 2006 | Corbesier, L., Vincent, C., Searle, I., Fomara, F., & Coupland, G. (2006). Analysis of the expression pattern of FT protein during flowering. COMPARATIVE BIOCHEMISTRY AND PHYSIOLOGY A-MOLECULAR & INTEGRATIVE PHYSIOLOGY, 143(4), S166. |
| 2006 | SEARLE, I. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev., 20, 898-912. |
| 2006 | SEARLE, I. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev, 20, 898-912. |
| 2006 | SEARLE, I. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev., 20, 898-912. |
| 2005 | Berendzen, K., Searle, I., Ravenscroft, D., Koncz, C., Batschauer, A., Coupland, G., . . . Ülker, B. (2005). A rapid and versatile combined DNA/RNA extraction protocol and its application to the analysis of a novel DNA marker set polymorphic between Arabidopsis thaliana ecotypes Col-0 and Landsberg erecta. Plant Methods, 1(1), 15 pages. Scopus68 WoS62 Europe PMC56 |
| 2004 | Searle, I., & Coupland, G. (2004). Induction of flowering by seasonal changes in photoperiod. EMBO Journal, 23(6), 1217-1222. Scopus238 WoS220 Europe PMC160 |
| 2003 | Searle, I. R., Men, A. E., Laniya, T. S., Buzas, D. M., Iturbe-Ormaetxe, I., Carroll, B. J., & Gresshoff, P. M. (2003). Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science, 299(5603), 109-112. Scopus472 WoS447 Europe PMC361 |
| 2003 | Waldron, J., Peace, C. P., Searle, I. R., Furtado, A., Wade, N., Findlay, I., . . . Carroll, B. J. (2003). Randomly Amplified DNA Fingerprinting: A culmination of DNA marker technologies based on arbitrarily-primed PCR amplification. Journal of Biomedicine and Biotechnology, 2(3), 141-150. Scopus22 Europe PMC16 |
| 2003 | SEARLE, I. R. (2003). Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science, 299, 109-112. |
| 2003 | SEARLE, I. R. (2003). Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science, 299, 109-112. |
| 2002 | May, K. J., Whisson, S. C., Zwart, R. S., Searle, I. R., Irwin, J. A. G., Maclean, D. J., . . . Drenth, A. (2002). Inheritance and mapping of 11 avirulence genes in Phytophthora sojae. Fungal Genetics and Biology, 37(1), 1-12. Scopus54 WoS45 Europe PMC42 |
| 2002 | Waldron, J., Peace, C. P., Searle, I. R., Furtado, A., Wade, N., Findlay, I., . . . Carroll, B. J. (2002). Randomly amplified DNA fingerprinting: A culmination of DNA marker technologies based on arbitrarily-primed PCR amplification. Journal of Biomedicine and Biotechnology, 2002(3), 141-150. Scopus18 |
| 2001 | SEARLE, I. R. (2001). Unknown Title. Towards the map-based cloning of nodulation genes from soybean. |
| 1999 | Schenk, G., Ge, Y., Carrington, L. E., Wynne, C. J., Searle, I. R., Carroll, B. J., . . . De Jersey, J. (1999). Binuclear metal centers in plant purple acid phosphatases: Fe-Zn in sweet potato and Fe-Zn in soybean. Archives of Biochemistry and Biophysics, 370(2), 183-189. Scopus151 WoS139 Europe PMC98 |
| Year | Citation |
|---|---|
| 2021 | Li, J., Wu, X., Do, T., Nguyen, V., Zhao, J., Ng, P. Q., . . . Searle, I. (2021). Quantitative and Single-Nucleotide Resolution Profiling of RNA 5-Methylcytosine. In Methods in Molecular Biology (Vol. 2298, pp. 135-151). Springer US. DOI Scopus1 |
| 2019 | Do, T., Qu, Z., & Searle, I. R. (2019). Purification and functional analysis of plant long noncoding RNAs (lncRNA). In J. Chekanova, & H. -L. Wang (Eds.), Plant long non-coding RNAs: methods and protocols (Vol. 1933, pp. 131-147). New York: Springer. DOI Scopus5 Europe PMC6 |
| Year | Citation |
|---|---|
| 2017 | Yang, S. Y., Tate, M., Asenstorfer, R., Carragher, J., Paull, J., & Searle, I. (2017). Progress Towards Developing Low Toxin Common Vetch (Vicia sativa L.). In BIT’s 5th World Congress of Agriculture-2017. Shenyang, China. |
| 2007 | Searle, I. R., Mosher, R. A., Melnyk, C. W., & Baulcombe, D. C. (2007). Small RNAs hit the big time: Meetings. In New Phytologist Vol. 174 (pp. 479-482). England: WILEY. DOI |
| Year | Citation |
|---|---|
| 2016 | Liu, H., Able, A., Searle, I., & Able, J. (2016). Characterising water deficit stress-responsive microRNAs and their functional targets in different durum wheat genotypes. Poster session presented at the meeting of ComBio 2016. Brisbane, Australia.. |
| 2015 | Liu, H., Searle, I., Watson-Haigh, N., Baumann, U., Mather, D., Able, A., & Able, J. (2015). Identification of differentially expressed miRNAs and their targets in durum wheat during water deficit stress. Poster session presented at the meeting of 9th International Wheat Conference. Sydney, Australia.. |
| 2015 | David, R., Lim, H. M., & Searle, I. R. (2015). Identification of pla/ubp14, an Arabidopsis mutant that displays lengthened plastochron and larger organs. Poster session presented at the meeting of Annual Genetics Society of Australasia conference. |
| Year | Citation |
|---|---|
| 2016 | Liu, H. (2016). Genome-wide characterisation of microRNAs and their target genes in different durum wheat genotypes under water limiting conditions. (PhD Thesis, University of Adelaide). |
| Year | Citation |
|---|---|
| 2015 | Liu, H., Searle, I., Watson-Haigh., Baumann., Mather, D. E., Able, A. J., & Able, J. A. (2015). Genome-wide identification of differentially expressed microRNAs in leaves and the developing head of four durum genotypes during water deficit stress. |
| Year | Citation |
|---|---|
| 2025 | Riley, S., Nguyen, V., Bhattacharjee, R., Ng, P. Q., Ritchie, T., Fisk, I., . . . Searle, I. (2025). TMT-based quantitative proteomic assessment of <i>Vicia sativa</i> induced neurotoxicity by β-cyano-L-alanine and γ-glutamyl-β-cyano-L-alanine in SH-SY5Y cells. DOI |
| 2025 | Xi, H., Llamas, B., Nguyen, V., Li, M., Zhou, Q., Liu, Z., & Searle, I. (2025). Two routes from the Fertile Crescent led to the introduction of common vetch into Europe. DOI |
| 2025 | Khanduja, J., Wu, X., Li, J., & Searle, I. (2025). The <i>Arabidopsis</i> CYSTM α 5’UTR increases protein production from transgenes in plants and bacteria. DOI |
| 2025 | Safdar, L. B., Fisk, I. D., Pasquariello, M., Lale, A., Searle, I. R., Bhosale, R. A., . . . Boden, S. A. (2025). Mutations in <i>HOMEOBOX DOMAIN-2</i> improve grain protein content in wheat without significantly affecting grain yield and senescence. DOI |
| 2024 | Wu, X., Keith Bullard, G., & Searle, I. (2024). A Quantitative Bioassay for Crown-Gall Tumourigenesis using Carrot Disks v1. DOI |
| 2018 | Xu, K., Zhang, X. -M., Yu, G., Lu, M., Liu, C., Chen, H., . . . Fu, Y. -F. (2018). <i>FT/FD-GRF5</i> repression loop directs growth to increase soybean yield. DOI |
- Competitive funding only listed below (since 2014)
- ARC Linage project-Chief Investigators Searle et al., Industry partner SAGIT (2021-2025)
- Yitpi foundation grant- Chief Investigators Searle and Nguyen (2020)
- SAGIT grant- Investigators Searle and Nagel (2020-2023)
- ARC Discovery Grant DP190101303- Investigators Searle and Studholme (2019-2021)
- Hermon Slade Foundation- Investigator Searle (2017-2020)
- ACSRF Australia-China Joint Research Centre of Grains for Health- Investigators, Searle, et al (2016-2019)
- ARC Future Fellowship FT130100525- Investigator Searle (2014-2018)
- ASTE Travel Grant- Investigator Searle (2015)
- ACSRF Australia-China Young Scientists Exchange Programme- Investigator Searle (2014)
- Interim UOC-UA Biotechnology program coordinator
- BIOTECH 7040- Coordinator for Applications of Next Generation Sequencing
- GENETICS 2510- Genetics IIA
- GENETICS 3211- Gene Expression & Human Developmental Genetics III
- SCIENCE 3500- Opening Doors to the Middle Kingdom
| Date | Role | Research Topic | Program | Degree Type | Student Load | Student Name |
|---|---|---|---|---|---|---|
| 2024 | Principal Supervisor | Role of an allele involving in stamen orientation of oilseed plant flowers that causes higher seed yield | Doctor of Philosophy | Doctorate | Full Time | Mr Jun Ming Loke |
| 2024 | Principal Supervisor | Role of an allele involving in stamen orientation of oilseed plant flowers that causes higher seed yield | Doctor of Philosophy | Doctorate | Full Time | Mr Jun Ming Loke |
| 2023 | Co-Supervisor | Characterising Physiological and Biochemical Traits in Shiraz Grapevines of Varying Vine Age in Barossa Valley Geographical Indication | Doctor of Philosophy | Doctorate | Full Time | Ms Grace Puspita Prayogo |
| 2023 | Co-Supervisor | Characterising Physiological and Biochemical Traits in Shiraz Grapevines of Varying Vine Age in Barossa Valley Geographical Indication | Doctor of Philosophy | Doctorate | Full Time | Ms Grace Puspita Prayogo |
| 2022 | Principal Supervisor | Does RNA m5C Regulate Post-Transcriptional Gene Expression in Plants? | Doctor of Philosophy | Doctorate | Full Time | Mr Jasjyot Singh Khanduja |
| 2022 | Principal Supervisor | Development of drought tolerant, high protein legume for arid Australia | Doctor of Philosophy | Doctorate | Full Time | Ms Junyi Wang |
| 2022 | Principal Supervisor | Development of drought tolerant, high protein legume for arid Australia | Doctor of Philosophy | Doctorate | Full Time | Ms Junyi Wang |
| 2022 | Principal Supervisor | Does RNA m5C Regulate Post-Transcriptional Gene Expression in Plants? | Doctor of Philosophy | Doctorate | Full Time | Mr Jasjyot Singh Khanduja |
| Date | Role | Research Topic | Program | Degree Type | Student Load | Student Name |
|---|---|---|---|---|---|---|
| 2020 - 2024 | Principal Supervisor | Construction of the Vicia sativa (common vetch) reference genome and exploration of genetic resources | Doctor of Philosophy | Doctorate | Full Time | Mr Hangwei Xi |
| 2020 - 2025 | Co-Supervisor | Advancing understanding of the genetic basis of grain protein content in wheat through hyperspectral imaging and genomics | Doctor of Philosophy under a Jointly-awarded Degree Agreement with | Doctorate | Full Time | Mr Luqman Bin Safdar |
| 2019 - 2021 | Principal Supervisor | The role of RNA modifications in Arabidopsis thaliana growth and development | Doctor of Philosophy | Doctorate | Full Time | Dr Pei Qin Ng |
| 2019 - 2023 | Principal Supervisor | Vetch as a New Protein Source for the Human Diet | Doctor of Philosophy under a Jointly-awarded Degree Agreement with | Doctorate | Full Time | Mr Samuel Arthur George Riley |
| 2018 - 2021 | Principal Supervisor | The Genetic Basis of Stamen Orientation - An Outcrossing Mechanism in the Brassicaceae | Master of Philosophy | Master | Full Time | Mrs Jayashrini Sakunthala Madawala |
| 2018 - 2022 | Principal Supervisor | Towards site-specific functional analysis of RNA N6-methyladenosine and 5-methylcytosine in Arabidopsis thaliana | Doctor of Philosophy | Doctorate | Full Time | Miss Huong Thi Thuy Ta |
| 2017 - 2021 | Co-Supervisor | Development and application of genomic approaches for resolving complex traits among insect pests | Doctor of Philosophy | Doctorate | Full Time | Mr Christopher Michael Ward |
| 2017 - 2021 | Principal Supervisor | Determining the molecular basis of beta-cyano-alanine and gamma-glutamyl-beta-cyano-alanine toxin accumulation in Vicia sativa | Doctor of Philosophy | Doctorate | Full Time | Dr Vy Hoang Thao Nguyen |
| 2017 - 2023 | Principal Supervisor | Characterizing RNA 5-methylcytosine in A. thaliana: Functions and Regulation Pathways | Doctor of Philosophy | Doctorate | Full Time | Miss Xingyu Wu |
| 2016 - 2019 | Co-Supervisor | Don't Cry for Me: Evolutionary and Functional Analysis of Two Rice Pollen Allergens, Ory s 1 and Ory s 12 | Doctor of Philosophy | Doctorate | Full Time | Ms Deborah Devis |
| 2016 - 2020 | Principal Supervisor | Identification and functional analysis of RNA modifications in Arabidopsis thaliana and Mus musculus | Doctor of Philosophy | Doctorate | Full Time | Miss Jun Li |
| 2016 - 2023 | Principal Supervisor | Investigating the role of RNA 5-methylcytosine on ribosomal and transfer RNAs in Arabidopsis thaliana | Doctor of Philosophy | Doctorate | Full Time | Miss Jing Zhao |
| 2014 - 2018 | Principal Supervisor | Identification and Functional Characterization of Long Noncoding RNAs Involved in Endosperm Development of Arabidopsis thaliana | Doctor of Philosophy | Doctorate | Full Time | Mr Quang Trung Do |
| 2013 - 2016 | Co-Supervisor | Genome-wide Characterisation of microRNAs and their target genes in different durum wheat genotypes under water limiting conditions | Doctor of Philosophy | Doctorate | Full Time | Dr Haipei Liu |
| 2013 - 2016 | Principal Supervisor | Conservation and Function of RNA 5-methylcytosine in Plants | Doctor of Philosophy | Doctorate | Full Time | Miss Alice Louise Burgess |
| Date | Role | Editorial Board Name | Institution | Country |
|---|---|---|---|---|
| 2011 - ongoing | Consulting Editor | Frontiers in Genetics and Genomics | - | - |
| Date | Office Name | Institution | Country |
|---|---|---|---|
| 2018 - ongoing | Australian Epigenetic Alliance- South Australian Representative | Australian Epigenetic Alliance (AEpiA) | Australia |
| 2016 - 2017 | Deputy Head of the Department of Genetics and Evolution | School of Biological Sciences, The University of Adelaide | Australia |
| 2015 - ongoing | GSA South Australian Representative on Executive | Genetics Society of AustralAsia | Australia |




